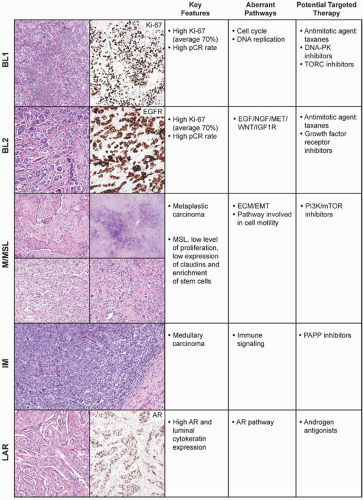
The pioneering molecular classification system for breast carcinoma was developed by Perou et al.
1 in 2000. This group performed cDNA microarray analysis on 65 samples from 42 patients. Therein, hierarchical data revealed distinctly variable gene expression patterns in different tumors, but similar patterns among paired samples of the same tumor. The “intrinsic” gene expression pattern was characteristic of an individual tumor—as opposed to those that varied as a function of tissue sampling. In this study, four molecular subtypes of breast carcinomas were identified: “luminal,” “HER2-enriched”, “basal-like”, and “normal breast-like”. The “
luminal group” of carcinomas were largely hormone receptor (1) and express luminal epithelial genes—traits similar to those of normal luminal epithelial cells. The “HER2-enriched group” was mainly composed of breast car cinomas with amplification of the HER2 gene. The “basal-like group” was
ER(-), and frequently corresponded to the triple negative breast carcinomas (TNBCs, i.e.,
ER(-),
PR(-), and HER2(-)). This group of tumors was immunoreactive for cytokeratin (
CK) 5/6 and CK17, similar to the reactivity pattern observed in myoepithelial cells of the normal breast epithelium (i.e., “basal” cells, hence the term applied to this group). The category of tumors called “normal breast-like” had a gene expression pattern similar to that observed in normal breast tissue. It subsequently became evident that the latter group most likely resulted from contamination of tissue samples with high levels of normal breast tissue, and may not exist at all.
ray analyses on 78 breast carcinoma samples. The results thereof further refined the intrinsic molecular classifier and proposed the division of
ER(+) luminal group of breast carcinomas into “luminal A” and “luminal B” subgroups. Although both groups expressed hormone receptors, the luminal B subgroup had a higher proliferation rate and lesser expression of
ER-related genes relative to those observed in luminal A group of tumors. Some luminal B tumors overexpressed the
HER2/neu gene. Luminal B tumors responded less to hormonal therapy, and more to chemotherapy, relative to luminal A tumors. In general, luminal B tumors had a poorer prognosis than did luminal A tumors. The “HER2-enriched” subtype was characterized by high expression of ERBB2 and genes in the ERBB2 amplicon at 17q22.24, including GRB7. Among HER2-overexpressed/amplified tumors, a significant proportion (50% or so) were
ER(+), and
ER(+)/HER2(+) tumors were classified as luminal B sub-group by both intrinsic classification systems. Additional studies also suggested that
ER(+) and HER2(+) (luminal B) breast carcinomas had high levels of genes often expressed
by normal luminal epithelial cells, whereas
ER(—)/HER2(1) (HER2-enriched) carcinomas had high levels of genes expressed by progenitor and stem cell-like cells.
3,4 These studies suggested that
ER(1)/HER2(1) (luminal B) and
ER(—)/ HER2(1) (HER2-enriched) breast carcinomas were biologically distinct. These two groups of tumors not only showed distinct patterns of responses to chemotherapy and HER2-targeted therapy but also displayed relatively distinctive time to and sites of relapse.
ER(—)/HER2(1) tumors tended to recur in the first 5 years, and recurrence after 5 years was low. In contrast,
ER(1)/HER2(1) tumors had lower recurrence rate during the first 5 years, but their tendency for recurrence persisted for 15 or more years.
ER(—)/HER2(1) tumors recurred more often in visceral organs, whereas
ER(1)/HER2(1) tumors recurred more often in bone. The latter pattern was also observed in
ER(1) luminal-type tumors. In addition,
ER(1)/HER2(1) tumors had a lower rate of pathologic complete response (
pCR) after neoadjuvant chemotherapy with or without trastuzumab than did
ER(—)/HER2(1) tumors, but
ER(1)/HER2(1) tumors did not seem to have poorer prognosis, compared with
ER(—) / HER2(1) tumors.
5,6 The clinical, pathologic, and molecular features of the basic molecular subtypes of breast carcinomas are summarized in
Table 45.1.