Fig. 1.1
Human pachytene spermatocyte demonstrating synapsis of homologous chromosome pairs and sites of meiotic recombination (chiasmata). The protein complex (synaptonemal complex) that forms the physical connection between homologous chromosomes is labeled in red, chromosome centromeres in blue, and sites of recombination in yellow. Note, all bivalents possess multiple chiasmata, with the exception of chromosomes 21 and 22 (arrowed) and the sex body which only possess a single chiasma; thus, these particular bivalents have an increased risk of chromosome nondisjunction if recombination does not take place (achiasmate bivalents)
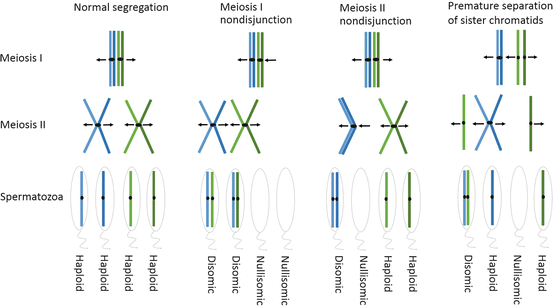
Fig. 1.2
Examples of nondisjunction mechanisms. The image on the left illustrates the normal segregation of homologous chromosomes in meiosis I and sister chromatids in meiosis II and the resulting spermatozoa. Following which, nondisjunction events occurring in meiosis I or meiosis II are shown, in which homologues (meiosis I) or sister chromatids (meiosis II) travel to the same pole. Note, in meiosis I the nondisjunction event may be due to a “true” nondisjunction event or the result of an achiasmate bivalent. The segregation pattern shown on the right illustrates how nondisjunction can arise as the result of premature separation of the sister chromatids (depicted in meiosis I, this can also arise in meiosis II). The outcome of these various segregation patterns will result in spermatozoa that will be either haploid (correct copy number), disomic (gain of a chromosome), or nullisomic (loss of a chromosome) for the chromosome(s) involved
1.6 Assessment of the Levels of Chromosome Aneuploidy Within Sperm
Given that the vast majority of chromosome aneuploidies perish in utero, often prior to a clinical pregnancy being established, an accurate assessment of the frequency can only be obtained by studying its frequency in the gametes (Templado et al. 2013). Since the advent of fluorescence in situ hybridization (FISH), chromosome aneuploidy has been relatively straightforward to assess within sperm (Fig. 1.3). FISH has been routinely utilized for decades in a wide range of clinical application; however, the majority of data on sperm aneuploidy is derived from research rather than clinical studies. To date, there are over 50 published studies examining the levels of sperm aneuploidy predominantly in normozoospermic and infertile men. These studies have revealed several important findings: (1) all men have a proportion of aneuploid sperm within their ejaculate, and (2) virtually all studies report significantly higher levels of aneuploidy in infertile men compared to individuals with normal semen parameters (Hann et al. 2011; Harton and Tempest 2012; Shi and Martin 2000; Tempest 2011; Tempest and Griffin 2004; Templado et al. 2011). From the published FISH studies, the lower estimates of sperm aneuploidy frequencies in males with normal semen parameters have been estimated to be around 3–5 % (Pang et al. 1999; Templado et al. 2011; Chatziparasidou et al. 2014). This aneuploidy estimate is based on extrapolated data given that only 2–5 chromosomes can be reliably scored in a single sperm cell due to the limited number of fluorochromes available. In addition, most studies have selected chromosomes that are clinically significant, namely, those chromosomes that are viable in an aneuploid state (chromosomes 13, 18, 21, X, and Y). For the most part, studies report similar levels of disomy with an average of approximately 0.1 % for each chromosome (Shi and Martin 2000; Tempest and Griffin 2004; Templado et al. 2011). These estimates are extrapolated from the data on investigated chromosomes; it is important to note that there are a number of chromosomes for which there is little or no aneuploidy FISH data available (Shi and Martin 2000; Tempest and Griffin 2004; Templado et al. 2011). However, the vast majority of studies have observed that specific chromosomes are more prone to chromosome nondisjunction with much higher frequencies of disomy found within sperm. Aneuploidy for chromosomes 21 and 22 and the sex chromosomes is frequently reported to be approximately threefold higher than the other chromosomes (Shi and Martin 2000; Tempest and Griffin 2004; Templado et al. 2011). In males, there is an average of 50 sites of recombination (chiasmata) in each pachytene spermatocyte; the chiasmata are distributed across chromosomes, and in the normal situation there is at least one chiasma located on the long and short arm of the chromosome (Hassold et al. 2000). Longer chromosomes such as chromosome 1 have a greater number of chiasmata (~5), whereas the smallest chromosomes 21 and 22 and the sex chromosomes usually only possess a single chiasma (Martin 2008; Sun et al. 2005, 2008). As discussed previously, meiotic recombination appears to play an important role in ensuring chromosomes disjoin correctly during meiosis. Therefore, the presence of multiple chiasmata along the length of a chromosome may function in part, as an insurance policy to prevent nondisjunction. Thus, smaller chromosomes possessing a single chiasma (e.g., chromosomes 21, 22, X, and Y) are more prone to be achiasmate (lacking a chiasma) (Sun et al. 2008), losing the insurance policy of additional chiasmata to reduce the risk of nondisjunction. Studies have demonstrated that some individuals have a higher proportion of achiasmate bivalents and that this translates to a significantly higher level of sperm aneuploidy (Ferguson et al. 2007; Sun et al. 2008).
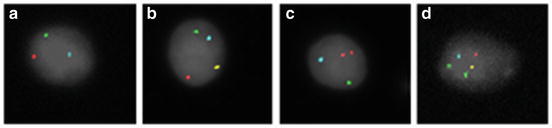

Fig. 1.3
Fluorescence in situ hybridization (FISH) for chromosomes 18, 21, X, and Y in sperm nuclei. The sperm nuclei are counterstained with DAPI (pseudo-colored gray) with chromosomes 18, 21, X, and Y probed in aqua, green, gold, and red fluorochromes, respectively. Panels (a–d) provide examples of normal and abnormal sperm FISH results for the investigated chromosomes: Panel (a) normal haploid Y-bearing sperm, Panel (b) XY disomy, Panel (c) YY disomy, and Panel (d) XY disomy and disomy 21
The vast majority of sperm FISH studies have analyzed and compared the frequencies of sperm aneuploidy between infertile men and either normozoospermic men, or men with proven fertility. All but a handful of these studies have identified a significant increase in sperm aneuploidy levels for at least one investigated chromosome in men with reduced semen parameters compared to normozoospermic men (Shi and Martin 2000; Tempest et al. 2004; Templado et al. 2011; Chatziparasidou et al. 2014). Collectively these studies provide strong evidence that infertile men have a significantly higher proportion of aneuploid sperm (approximately threefold), with severe oligoasthenoteratozoospermic and nonobstructive azoospermic individuals often having the highest levels (Shi and Martin 2000; Tempest et al. 2004; Templado et al. 2011; Chatziparasidou et al. 2014). From the published data, it is also clear that there are notable differences in the frequencies reported between studies; thus, baseline levels are difficult to establish. Differences between studies are likely due to technical differences including FISH probes utilized, number of cells scored, differences in scoring criteria, and subjective differences between scorers. In addition, the differences could be due to interindividual differences in sperm aneuploidy frequencies. It would seem plausible that some individuals may be more prone to chromosome nondisjunction and anaphase lag and/or have a less efficient meiotic checkpoint than others that may lead to increased levels of sperm aneuploidy. Furthermore, exogenous factors may have the ability to increase or decrease sperm aneuploidy levels. To date, several studies have examined whether sperm aneuploidy levels are consistent over time and if interindividual differences in sperm aneuploidy exist. The general consensus of these studies reveals that sperm aneuploidy levels remain remarkably consistent over time within individuals; nevertheless, there are stable variants who consistently produce higher levels of sperm aneuploidy (Rubes et al. 2005; Tempest et al. 2009). Preliminary data suggests that exogenous factors (e.g., diet, chemotherapy, or environment) may affect sperm aneuploidy levels resulting in a transient increase or potentially a decrease in aneuploidy levels (Harkonen 2005; McAuliffe et al. 2012, 2014; Tempest et al. 2005, 2009; Young et al. 2013). Despite the findings of significantly higher levels of chromosome aneuploidy within the sperm or certain subsets of men, sperm aneuploidy is rarely examined clinically, with the test only available at a handful of reference laboratories worldwide (Carrell 2008; Ramasamy et al. 2014). The lack of robust clinical studies provides us with a relatively poor understanding of the role of sperm aneuploidy in embryogenesis. Given that certain individuals produce higher levels of sperm aneuploidy, perhaps the most important question to address is whether these increased levels translate to an increased risk of paternally derived aneuploid embryos and offspring. This question is particularly difficult to address and is confounded by the significant maternal contribution to chromosome aneuploidy and that the parental origin of trisomies is rarely identified. Thus, if high sperm aneuploidy levels are identified, it is not yet clear how, or if, this should be utilized to counsel patients (Harton and Tempest 2012; Carrell 2008; Hann et al. 2011; Tempest 2011; Templado et al. 2011). To date, a handful of studies have tried to address whether sperm aneuploidy levels translate to embryo aneuploidies. Several studies have retrospectively identified increased levels of sperm aneuploidy in the fathers of paternally derived aneuploid offspring (Arnedo et al. 2006; Blanco et al. 1998; Martinez-Pasarell et al. 1999; Soares et al. 2001; Moosani et al. 1999). These studies suggest that in almost all cases these individuals had significantly higher levels of sperm aneuploidy for multiple chromosomes compared to fertile men with no history of aneuploid offspring (Harton and Tempest 2012). Other studies have provided preliminary evidence to suggest that higher levels of sperm aneuploidy are associated with recurrent ICSI failure (Nicopoullos et al. 2008; Petit et al. 2005), increased chromosome abnormalities in preimplantation embryos (Gianaroli et al. 2005), and lower pregnancy rates and live births (Nagvenkar et al. 2005). Furthermore, the approximate threefold increase in sperm aneuploidy observed in infertile men is mirrored by a threefold increase in de novo chromosome aberrations observed in children born after ICSI (Aboulghar et al. 2001; Bonduelle et al. 2002; Devroey and Van Steirteghem 2004; Van Steirteghem et al. 2002). Clearly, none of these studies provide direct evidence that sperm aneuploidy directly translates to embryo aneuploidy. However, albeit in a very small number of studies, there is compelling evidence to suggest that sperm aneuploidy may play a greater role in transmitting aneuploidy to embryos than previously perceived (Harton and Tempest 2012; Tempest 2011).
1.7 How Do Chromosome Aberrations Affect Meiosis?
1.7.1 Numerical Sex Chromosome Aneuploidies and Their Impact on Fertility and Meiosis
Sex chromosome aneuploidies are relatively common in the general population. The incidence of both Klinefelter syndrome (47,XXY) and 47,XYY syndrome is estimated to occur in 1 in 500 to 1 in 1,000 male live births (Morris et al. 2008). Individuals with Klinefelter syndrome typically present with nonobstructive azoospermia (accounting for ~11 % of nonobstructive azoospermia) or potentially oligozoospermia with a mosaic karyotype (Van Assche et al. 1996). Klinefelter syndrome is often perceived to have a classical phenotype (e.g., tall stature, gynecomastia, and hypogonadism); however, many cases have a highly variable phenotype and may not be identified until they present with infertility (Paduch et al. 2009). These men are born with spermatogonia, but during early puberty the spermatogonia undergo a massive wave of apoptosis leading to azoospermia. In around 50 % of cases, sperm can be recovered following testicular sperm extraction and can be used with ICSI to allow patients to have biological offspring (Paduch et al. 2009). Individuals with a 47,XYY karyotype also present with a variable phenotype; the majority of cases may have no phenotypic abnormalities, whereas some individuals may have a greater risk for behavioral problems, mild learning disabilities, and tall stature (Kim et al. 2013). Men with 47,XYY syndrome exhibit variable semen parameters ranging from normozoospermia to azoospermia (Kim et al. 2013) and hence, as with Klinefelter syndrome, may only be diagnosed if they present with fertility problems. In both of these cases, there is an additional sex chromosome that has to proceed and be segregated through meiosis; therefore, if sperm is present, there is a theoretical risk that 50 % of the sperm produced will be aneuploid for the sex chromosomes. Thus, in these situations there is a significant risk of fetal demise and transmission of a sex chromosome aneuploidy in future offspring. FISH has been utilized to assess the levels of sex chromosome aneuploidy in the sperm from 47,XXY and 47,XYY males. The published studies are highly variable, but report significantly lower sperm aneuploidy levels than the theoretical 50 %. In brief, the range of sperm aneuploidy for the sex chromosomes reported in Klinefelter syndrome (non-mosaic/mosaic) and 47,XYY men is between 1–25 %/0–7 % and 0.1–14 %, respectively (Ferlin et al. 2005; Sarrate et al. 2005; Tempest 2011). These studies provide additional evidence of the presence of an as-of-yet unknown meiotic checkpoint that is relatively efficient in eliminating a large proportion of aneuploid sperm cells (Harton and Tempest 2012; Burgoyne et al. 2009) and may contribute to the low sperm count observed in some 47,XYY males. Nonetheless, despite the presence of a meiotic checkpoint, there remains a considerable proportion of aneuploid sperm that are capable of completing meiosis leading to significantly higher levels of sperm aneuploidy compared to karyotypically normal men. To date, several studies have reported that the increase in sperm aneuploidy observed is also mirrored by an equivalent increase in sex chromosome aneuploidies in preimplantation embryos (Gonzalez-Merino et al. 2007; Staessen et al. 2003). Additionally, approximately 10 % of cases in the literature have resulted in aneuploid offspring (two 47,XXY conceptuses) (Friedler et al. 2001; Ron-El et al. 2000). Therefore, couples should be offered genetic counseling to inform them of their potential increased risk of spontaneous abortions and aneuploid offspring.
1.7.2 Structural Alterations in the Paternal Genome and Their Impact on Fertility and Meiosis
Structural chromosome aberrations can be classified as cytogenetic aberrations or genomic structural variants and can include translocations, inversions, insertions, or deletions. Chromosome aberrations are considered balanced when all the DNA is present, but its order or location on a chromosome(s) has been rearranged or unbalanced if there is gain or loss of genetic material. Cytogenetically visible aberrations can be readily detected by karyotyping and are relatively large in size (>3–5 Mb) (Shaffer and Bejjani 2004), whereas genomic structural variants are not microscopically visible (typically, >1 kb) (Freeman et al. 2006).
1.7.3 Karyotype Aberrations and Their Impact on Fertility and Meiosis
The incidence of cytogenetically visible chromosome aberrations is higher in males with fertility problems than that of the general population (Harton and Tempest 2012). Carriers of balanced structural chromosome rearrangements (e.g., translocations and inversions) are usually phenotypically normal; however, they often present with infertility. Structural chromosome rearrangements pose problems during meiosis, requiring unique pairing structures to form and to facilitate homologous chromosome pairing (e.g., quadrivalents, trivalents and inversion loops for reciprocal translocations, Robertsonian translocations and inversions, respectively). The formation of these unique pairing structures imposes time constraints and/or may lead to failure of chromosome pairing, which could result in the activation of meiotic checkpoints potentially eliminating these cells (Shah et al. 2003). However, some cells are able to successfully form these unique pairing structures and complete meiosis. In this situation, problems can arise when segregating chromosomes to daughter cells.
1.7.4 The Impact of Chromosome Translocations on Fertility and Meiosis
In the case of chromosome translocations, several outcomes are possible: gametes may (1) contain the normal chromosome complement, (2) be balanced carrying the chromosome translocation, or (3) be unbalanced (containing segments that may be monosomic and/or trisomic for the chromosome regions involved in the translocation). Sperm cells that are chromosomally normal or balanced are unlikely to affect embryogenesis or development. However, a significant proportion of sperm will be unbalanced. The phenotypic consequences of unbalanced segregation in chromosome translocations are often difficult to predict, given that most chromosomes translocations are unique to individual families. In these cases a careful review of the family history and assessment of the chromosomes involved and the size of the translocation can assist with counseling for the various possible outcomes. The vast majority of unbalanced sperm would result in early embryonic arrest or spontaneous abortions due to the incompatibility of partial trisomies and monosomies with embryogenesis. However, it is important to note that some unbalanced segregation products may be clinically viable, potentially resulting in congenital malformations and cognitive impairment. This has an increased likelihood if the chromosome translocation involves gene-poor regions of the genome, is small in size, and/or is known to be tolerated in a monosomic (e.g., chromosome X) or trisomic state (e.g., chromosomes 13, 18, 21, X, and Y). Using FISH probes for the specific chromosomes involved in the translocation, it is possible to evaluate the proportion of unbalanced sperm produced. The published literature reports extremely variable levels of sperm aneuploidy, which likely reflects patient-specific translocations. Studies evaluating the frequencies of unbalanced sperm from 30 reciprocal translocation carriers report partial aneuploidy levels of 29–81 % (Ferlin et al. 2007; Sarrate et al. 2005; Tempest 2011), whereas studies from 20 Robertsonian translocation carriers report aneuploidy levels of between 3 and 36 % (Ferlin et al. 2007; Sarrate et al. 2005; Tempest 2011).
1.7.5 The Impact of Chromosome Inversions on Fertility and Meiosis
Meiotic segregation in carriers of balanced inversions usually results in normal or balanced gametes, unless meiotic recombination takes place within the inversion. If recombination takes place within the inversion, resulting gametes may be monosomic and/or trisomic for the regions involved and potentially acentric or dicentric if the inversion involves the centromere (paracentric inversion). The clinical viability of unbalanced gametes is essentially the same as for translocations, in that, the vast majority will be spontaneously aborted often prior to a clinical pregnancy being established or potentially viable with risks of congenital abnormalities and/or cognitive impairment depending on the chromosome region involved and size of the unbalanced segments. In the case of inversions, larger inversions have a higher likelihood of resulting in unbalanced gametes as there is an increased risk of recombination occurring within a larger segment. To date, a handful of studies have examined the proportion of unbalanced sperm using FISH. As with translocations, the percentage of unbalanced sperm produced in inversion carriers varies widely between studies (1–54 %) (Anton et al. 2002, 2005; Jaarola et al. 1998; Mikhaail-Philips et al. 2004, 2005; Yakut et al. 2003), with the highest levels most likely reflecting larger inversions.
In the case of translocations and inversions, the estimates of sperm aneuploidy should be used with caution and are not generally applicable due to the fact that the vast majority of aberrations are unique. Therefore, individual assessment using personalized FISH probes for the chromosomal rearrangement can be readily used to obtain patient-specific risks in conjunction with family history to determine frequency of unbalanced sperm and whether unbalanced products may be clinically viable.
1.7.6 Genomic Structural Aberrations
It is clear that cytogenetically visible chromosome aberrations, numerical or structural in nature, can have a tremendous impact on embryogenesis resulting in embryonic arrest, fetal demise, and potentially live-born offspring with wide-ranging clinical phenotypes. Despite the clear association of abnormal karyotypes and embryogenesis, it is noteworthy that a significant proportion of early failed pregnancies with developmental defects are karyotypically normal. Studies suggest that up to ~20 % of morphologically abnormal embryos possess a normal karyotype (Philipp et al. 2003; Rajcan-Separovic et al. 2010). It seems plausible that many of these seemingly euploid developmentally abnormal embryos may be the result of submicroscopic perturbations that are not detectable by routine karyotyping. Recent technological advances that include chromosomal microarrays and sequencing now enable the routine detection of insertions, deletions, inversions, and duplications that are as small as 50 bp in size. These variants can arise through multiple mechanisms; however, they frequently arise as a result of errors during meiotic recombination. The paternal and maternal transmission of de novo submicroscopic chromosome aberrations following errors in meiotic recombination is likely to be equal, given that both male and female gametes only progress through meiosis on a single occasion. De novo submicroscopic chromosome aberrations can be benign or pathogenic in nature depending on their location and genetic content (Wapner et al. 2012). The clinical utilization of chromosome microarrays has been extremely beneficial in the evaluation of children with neurocognitive developmental delays and congenital structural malformations (Wapner et al. 2012). However, the consequence of these submicroscopic aberrations on fertilization, embryogenesis, and fetal development remains poorly understood. To date, a handful of studies have reported that between 4 and 13 % of miscarriages may possess submicroscopic chromosome aberrations that would not be detectable by routine karyotyping (Rajcan-Separovic et al. 2010; Shaffer et al. 2008; Shimokawa et al. 2006). It is important to note that the majority of studies did not determine whether these submicroscopic alterations were de novo, benign, or potentially pathogenic. A recent large study compared the use of chromosomal microarray and karyotyping for prenatal diagnosis enrolling over 4,400 women with indications for prenatal diagnosis including advanced maternal age, abnormal Down syndrome screening result, and structural abnormalities identified on ultrasound. This study reported that 6 % of samples were found to have a submicroscopic clinically significant unbalanced chromosome aberration (Wapner et al. 2012). The occurrence of submicroscopic aberrations in the prenatal diagnosis study is on the lower end compared to that reported in the miscarriage studies; this is most likely due to the fact that the prenatal diagnosis study evaluated gestationally older pregnancies than the miscarriage studies and only included potentially pathogenic alterations. The gestational age is critical when considering chromosome aberrations. As embryos develop a lower proportion of chromosomally abnormal embryos will be identified as the vast majority of aberrations result in spontaneous abortions, thus the prevalence of chromosome aberrations decreases with gestational age. Another critical point to note is the inability of chromosome microarrays used by these studies to detect balanced chromosome aberrations or triploidy; therefore, prevalence of the chromosomal aberrations reported could be conceivably higher (Wapner et al. 2012).
1.8 Conclusions
Chromosome aneuploidy, complete or partial in nature, can perturb embryogenesis and development. Somewhat surprisingly, the role of paternally derived chromosome aneuploidy in embryogenesis and development remains questionable in the clinic. A survey administered to fertility clinics in the UK perceived there is to be an increased risk of transmitting paternal genetic abnormalities following ICSI, but despite the concern most did not offer sperm aneuploidy screening (Griffin et al. 2003). Published studies suggest sperm aneuploidy has the potential to be a clinically useful screening tool to identify individuals with an increased risk of transmitting chromosome aneuploidy to future offspring. However, the clinical implementation of widespread routine sperm aneuploidy screening has been hampered by a number of factors that include (1) large maternal contribution to aneuploidy, (2) various technical challenges (e.g., wide variations between studies, requirement to score large numbers of cells (>5,000), limited number of chromosomes tested (3–5 chromosomes) per cell, and the inability to test the individual sperm that will be used for ICSI), (3) identification of individuals who may benefit from sperm aneuploidy scoring, and (4) what is a clinically significant level of sperm aneuploidy and how should patients with higher levels of aneuploidy be counseled. These important considerations have largely precluded the widespread clinical application of sperm aneuploidy screening, suggesting that at the moment the drawbacks outnumber the potential benefits. However, it is important to note that published studies have demonstrated chromosome aberrations (numerical or structural) can initiate and complete meiosis. Furthermore, the levels of aneuploidy observed in the sperm of infertile men or men with karyotype aberrations are mirrored and translate to increased levels of aneuploidy in preimplantation embryos and offspring, with several potential outcomes including fetal demise and congenital and cognitive impairments. Nevertheless, these findings are fundamentally based on a handful of small studies, and the true clinical ramifications of such findings will be difficult to determine until larger studies in a clinical setting are initiated. It seems unlikely that such studies will be initiated until some of the technical issues are resolved allowing rapid, cost-effective assessment of sperm aneuploidy levels. Automated capture and analysis software is currently available for FISH scoring and has been successfully applied to measure sperm aneuploidy and more commonly clinically for oncological FISH assays. However, automated approaches to assess sperm aneuploidy are not available in most clinics and have failed to significantly reduce the cost of the test (Carrell and Emery 2008; Tempest et al. 2010), primarily due to the initial outlay for capture system, software, and the relatively high cost of commercial FISH probes. Additionally, due to the lack of distinguishable fluorochromes, scoring all 24 chromosomes simultaneously in a single cell does not provide a genome-wide appreciation of the levels of aneuploidy within a single cell. The development and application of rapid multicolor FISH assays in combination with FISH reprobing and automation may allow all 24 chromosomes to be assessed in a single cell (Ioannou et al. 2011, 2012). Furthermore, it is also important to note that FISH as it is currently performed for aneuploidy assessments provides an extremely low resolution, typically only assessing a single chromosome region. Furthermore, FISH does not readily provide the possibility to screen for de novo structural aberrations or small genomic variants that may significantly contribute to spontaneous abortions. Technologically, it is possible to assess genome-wide levels of nondisjunction, unbalanced rearrangements, and smaller genomic variants in a single sperm cell using higher-resolution methodologies (e.g., sequencing, chromosome microarrays, and SNP arrays). Currently this remains prohibitively expensive as a routine clinical test given the large number of sperm cells that would need to be screened due to the relatively low proportion of aneuploidy in sperm. Currently, estimates of the proportion of sperm with de novo structural aberrations and smaller genomic perturbations are unknown, but it is reasonable to suggest that these may be similar or lower than aneuploidy levels. Thus, globally the proportion of sperm that may be perturbed may be clinically significant but still relatively low and hence not cost-effective to evaluate unless specific patient cohorts who may benefit can be identified.
Stay updated, free articles. Join our Telegram channel

Full access? Get Clinical Tree


