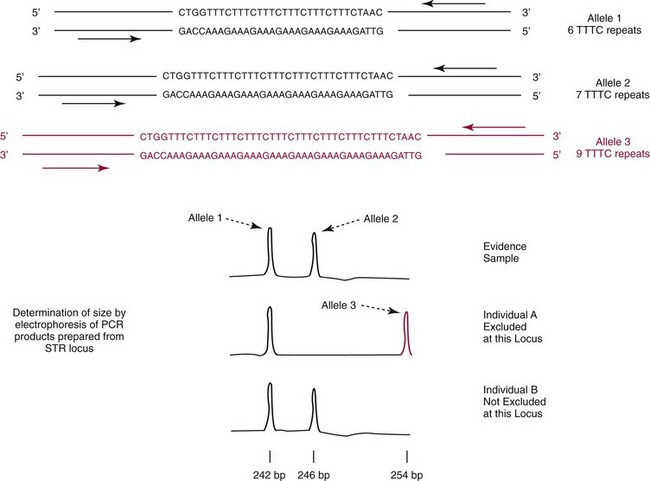
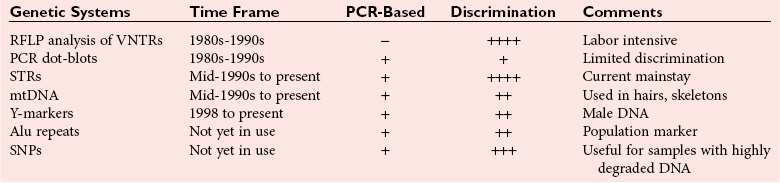
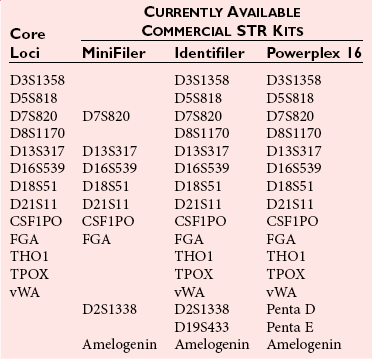
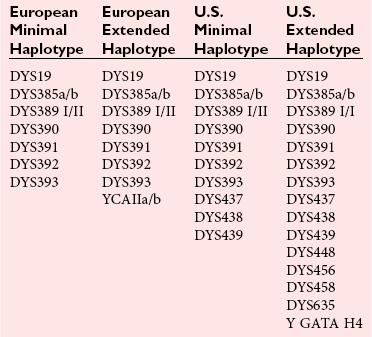
Chapter 41 Thomas M. Williams, M.D., Howard J. Baum, Ph.D., Victor W. Weedn, M.D., J.D. and Malek Kamoun, M.D., Ph.D. Several classes of genetic variants are found in the genome; some are more useful than others for identity testing. Most of the variants used occur in the noncoding genetic regions, such as introns, regulatory domains, and regions between genes, whereas some variants occur in gene domains transcribed into RNA, that is, the exons. Highly repetitive sequence elements that contribute to the structure of centromeres and telomeres and hundreds of thousands of copies of transposable elements that move about the genome over time may vary among individuals. However, these repetitive sequence elements generally are not useful for identity testing. Several million single-nucleotide polymorphisms (SNPs) have been identified in the genome (see Chapter 38). A subset of SNPs can be identified based on the ability of a restriction endonuclease to digest double-stranded DNA at the site of the variation. These SNPs are referred to as restriction fragment length polymorphisms (RFLPs). SNPs and most RFLPs are not very useful for identity testing because they usually have only two alleles. Short tandem repeat (STR) or microsatellite loci consist of DNA sequence motifs that have core repeats of two to seven base pairs.6,15 Examples include the dinucleotide 5′ CACACACA 3′ and the tetranucleotide 5′ TTTATTTATTTA 3′. Thousands of STRs are scattered throughout the genome. Because they are flanked by unique sequences, each can be specifically amplified with the polymerase chain reaction (PCR) for analysis. In populations of individuals, multiple alleles may be present based on differences in the numbers of repeated motifs at the locus. STRs have many characteristics that make them ideal for identity testing: (1) they can be analyzed in fluorescent automated systems; (2) alleles can be assigned in a definitive manner following analysis; (3) STR loci are almost always transmitted in families in a Mendelian fashion; (4) the loci may have 10 or more alleles, often with substantial allele frequencies, making them highly informative and making it easier to resolve mixtures of DNA; and (5) extensive information is available about allele frequencies in many human populations for STRs commonly used in identity testing.7 Commercially available STR systems employ tetrameric and pentameric repeat loci, which produce fewer artifactual bands and are characterized by roughly equal amplification of both alleles within an individual (Figure 41-1). Fragments can be labeled during PCR amplification with fluorescently tagged primers that facilitate multiplexing. DNA testing has revolutionized criminalistics.19,33 Only fingerprint evidence can sometimes rival the ability of DNA as trace evidence left at a scene to identify a perpetrator. As a general rule, other trace evidence merely links an article, instrument, or material to a scene. The origin of DNA-based identity testing is generally traced to a 1985 article in Nature by Alec Jeffreys.18 He coined the term “DNA fingerprint” and suggested that the hybridization of DNA probes to polymorphic genetic loci could be exploited for forensic purposes. Jeffreys first applied his techniques to civil and criminal cases in England. In the United States, DNA-based identity testing was introduced via commercial laboratories and later the Federal Bureau of Investigation (FBI). Today approximately 200 forensic DNA typing laboratories have been established in the United States, along with many other DNA laboratories around the globe. Forensic DNA testing is also used to identify decomposed unknown human remains through kinship analysis and can be used to identify victims of a mass disaster.4 Numerous genetic systems that are employed by forensic laboratories are summarized in Table 41-1.3,24 Jeffreys described a method to create a barcode-like DNA fingerprint based on VNTRs. Because the probes used bind to several loci in Southern hybridization experiments and produce numerous bands per probe, they are termed multilocus probes. In the United States, laboratories have preferred single-locus probe (SLP) systems that hybridize only to a single genetic locus. However, Southern hybridization–based RFLP analysis is expensive, labor intensive, difficult to automate, and less sensitive than PCR-based methods. Finally, RFLP is sensitive to DNA degradation—an important issue with environmentally exposed forensic specimens. RFLP tests have been largely abandoned in favor of the more efficient PCR-based assays discussed later.5 Most identity testing performed today relies on the PCR. PCR testing is inherently sensitive, allowing routine analysis of nanogram quantities of genomic DNA and often successful testing of picogram quantities (one cell contains 5 to 10 pg of DNA). Low copy number (LCN) STR analysis8 detects quantities of DNA down to the single cell level PCR, which underlies the characterization of STR and other loci for forensic identity testing loci described in this and later sections. The FBI laboratory’s combined DNA index system (CODIS) blends forensic science and computer technology into an effective tool for investigating violent crimes. CODIS enables federal, state, and local crime laboratories to exchange and compare DNA profiles electronically, thereby linking crimes to each other and to convicted offenders. The FBI convened a panel of forensic scientists in 1998 that chose a panel of 13 STR loci for use in the National DNA Index System. These 13 core loci have become the standard for casework and databanking for most forensic laboratories around the world (Table 41-2). They have been commercialized as kits in a variety of formats. STRs are now routinely used in crime laboratories globally and typically yield discriminatory values of one in trillions to sextillions. Y chromosome polymorphic loci can be used as identifying loci found only in males. In this way, male-specific DNA obtained from a vaginal swab can be typed without the usual differential extraction, in which the DNA from spermatozoa is released after the female fraction has been isolated from epithelial cells. Y chromosome loci useful for identity testing include STRs or SNPs (described later in this chapter). Laboratories typically employ commercially available panels of 12 to 17 Y chromosome STRs for analyses (Table 41-3). Y chromosome SNPs are in development. In the human mitochondrial genome, only approximately 1200 bases in the region of transcription origin (15971-579), known as the displacement loop (D-loop) or the control region, are noncoding. This D-loop consists of two hypervariable regions that contain the majority of polymorphisms useful for identity (HVI: 16024-16365; HVII: 73-340). Polymorphisms outside this region can also be employed for testing. mtDNA polymorphisms are typically identified for forensic testing via DNA sequencing of hypervariable regions.15 This method is expensive, labor intensive, and highly sensitive to contamination. The mtDNA sequence obtained from a specimen is compared with a reference sequence (revised Cambridge sequence, www.mitomap.org/MITOMAP, accessed May 24, 2010). As in the case with Y chromosome STRs, because mtDNA polymorphisms are linked, individual polymorphism frequencies cannot be multiplied together to generate a likelihood of identity such as by independently segregating chromosomal locus allele frequencies. Instead the mtDNA haplotype identified in a sample is compared with those deposited in a database to derive a frequency statistic. Many mitochondrial haplotypes in the database are unique. Because the database has more than 6000 entries, it can be fairly stated that many mitochondrial haplotypes have a discriminatory value greater than 1 in 6000. However, 18 common haplotypes have population frequencies greater than 0.5%, including a haplotype present in 7% of the population. In aggregate, these common haplotypes account for 20% of all haplotypes.
Identity Assessment
Variation in the Human Genome
Genetic Variation Useful in Identity Testing
Forensic Dna Typing
Genetic Systems Used in Forensic Identification
VNTR Analysis by RFLP
Short Tandem Repeats
Gender Markers and Y Chromosome Markers
Mitochondrial DNA
![]()
Stay updated, free articles. Join our Telegram channel

Full access? Get Clinical Tree


Basicmedical Key
Fastest Basicmedical Insight Engine