Herpesviridae
Philip E. Pellett
Bernard Roizman
Who made the world I cannot tell;
‘Tis made, and here I am in hell.
My hand, though now my knuckles bleed,
I never soiled with such a deed.
A. E. Housman, No. XIX in More Poems
Definition
Inclusion in the Family Herpesviridae
Herpesviruses have historically been defined based on the architecture of the virion (Fig. 59.1). A typical herpesvirion consists of a core containing a linear double-stranded DNA (dsDNA, ranging from 124–295 kb in length); an icosahedral capsid approximately 125 nm in diameter containing 161 capsomeres with a hole running down their long axis, plus one capsomeric structure that serves as the portal for packaging and release of the viral genome (the complex of the core and capsid is the nucleocapsid); an amorphous-appearing, sometimes asymmetric material that surrounds the nucleocapsid and is designated the tegument; and an envelope containing viral glycoprotein spikes on its surface. Based on the morphologic criteria, highly divergent viruses with hosts that range from bivalves to humans have been identified as herpesviruses (Fig. 59.2). Originally classified into a single family, the availability of extensive nucleotide sequence data led to establishment of a new taxonomic order, the Herpesvirales,28,102 that encompasses three virus families: the herpesviruses of mammals, birds, and reptiles (the Herpesviridae),100 herpesviruses of fish and amphibians (the Alloherpesviridae),99 and herpesviruses of bivalves (the Malacoherpesviridae).101 This and subsequent chapters in this book are concerned primarily with viruses that have long been recognized as the family Herpesviridae.
The objectives of this chapter are to provide definitions and examples of many of the terms and concepts that are relevant across the diverse collection of herpesviruses (a glossary is provided in e-Table 59.1). Briefly summarizing such a broad area of active research can be accomplished only at the cost of oversimplification and overgeneralization. Subsequent chapters will delve into most of these areas in much greater depth. Students, especially, should understand that the paradigms presented here represent significant opportunities for conceptual and experimental challenge.
Distribution in Nature
Herpesviruses are highly disseminated in nature. Most animal species have yielded at least one herpesvirus and frequently several distinct herpesviruses on examination. Inasmuch as few herpesviruses naturally infect more than one species, the number of herpesviruses in nature is likely to exceed the more than 200 identified to date. Thus far, nine herpesviruses have been identified that have humans as their primary host: herpes simplex virus 1 (HSV-1), herpes simplex virus 2 (HSV-2), human cytomegalovirus (HCMV), varicella-zoster virus (VZV), Epstein-Barr virus (EBV), and Human herpesviruses 6A, 6B, and 7 (HHV-6A, HHV-6B, HHV-7), and Kaposi’s sarcoma–associated herpesvirus (also known as HHV-8). Some key properties of many of the known herpesviruses are listed in Table 59.1, and a more comprehensive list of viruses is provided in e-Table 59.2. We have not tabulated the many herpesviruses identified primarily on the basis of small segments of nucleotide sequence (e.g., 35,36 and references therein).
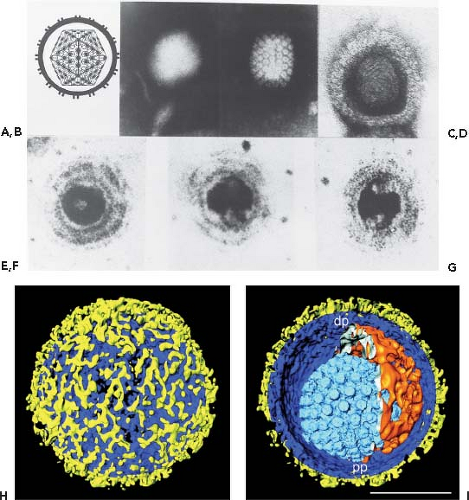 Figure 59.1. Herpesvirus morphology as visualized by transmission electron microscopy. A: Schematic representation of the herpesvirion seen through a cross-section of the envelope with spikes projecting from its surface. The sides of the icosahedron forming the capsid show twofold symmetry. The irregular inner perimeter of the envelope is meant to represent the occasional asymmetric arrangement of the tegument. B: An intact negatively stained HSV-1 virion. The intact envelope is not permeable to negative stain. The diameter of the virion is approximately 120 nm. C: A HSV-1 capsid exposed to negative stain and showing twofold symmetry matching the diagrammatic representation of the capsid in A. D: HSV-1 nucleocapsid containing DNA permeated with uranyl acetate. The electron micrograph shows the presence of thread-like structures 4 to 5 nm wide on the surface of the core. E–G: Electron micrographs of thin sections of HSV-1 virions showing the core cut at different angles. The preparation was stained with uranyl acetate and counterstained with lead citrate. The DNA core preferentially takes up the stain and appears as a toroid with an outer diameter of 70 nm and an inner diameter of 18 nm. The toroid appears to be suspended by a fibrous cylindrical structure. The micrographs show the toroid seen looking down the hole (E), in cross-section (F), or from the side (G). The electron micrographs shown in D to G are from Furlong et al.41 H, I: Segmented surface rendering of a single virion tomogram after denoising. Outer surface showing the distribution of glycoprotein spikes (yellow) protruding from the membrane (blue) (H). Cutaway view of the virion interior, showing the capsid (light blue) and the tegument “cap” (orange) inside the envelope (blue and yellow) (I). pp, proximal pole; dp, distal pole. Bar = 100 nm. (A, B, C, H, I from Grunewald K, Desai P, Winkler DC, et al. Three-dimensional structure of herpes simplex virus from cryo-electron tomography. Science 2003;302:1396-98, with permission. D through G from Furlong D, Swift H, Roizman B. Arrangement of herpesvirus deoxyribonucleic acid in the core. J Virol 1972;10:1071–1074, with permission.) |
Biological Properties
Members of the family Herpesviridae share four significant biological properties:
They specify a large array of enzymes involved in nucleic acid metabolism (e.g., thymidine kinase, thymidylate synthetase, dUTPase, ribonucleotide reductase), DNA synthesis (e.g., DNA polymerase, helicase, primase), and processing of proteins (e.g., protein kinases), although the exact array of enzymes may vary from one herpesvirus to another (Table 59.2).
Virus gene transcription, synthesis of viral DNA, and nucleocapsid assembly occur in the nucleus. Most virions
acquire at least part of their tegument and are enveloped in the cytoplasm.
Production of infectious progeny virus (lytic infection) is generally accompanied by the destruction of the infected cell.
The herpesviruses examined to date employ cellular latency as a mechanism for lifelong persistence in their hosts (Fig. 59.3).
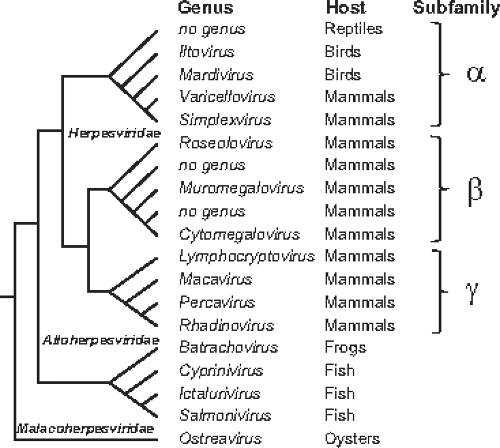 Figure 59.2. Major phylogenetic relationships and taxonomic subunits within the order Herpesvirales.102 The schematic shows branching patterns, not evolutionary distances. |
Herpesviruses also differ with respect to many of their biological properties. Some have a wide host cell range, multiply rapidly, and quickly destroy the cells they infect (e.g., HSV-1, HSV-2). Others have a narrow host cell range (EBV, HHV-6) or a long replicative cycle (HCMV). Herpesviruses can differ substantially with respect to the details of the mechanisms they use to manage host responses to infection and in the pathogenic mechanisms and clinical manifestations of diseases they cause.
Nomenclature and Classification
Herpesvirus Species
The definition of viral species accepted by the International Committee on Taxonomy of Viruses (ICTV) is “A virus species is a polythetic class of viruses that constitutes a replicating lineage and occupies a particular ecological niche”.125 Consistent with this, “A herpesvirus may be classified as a species if it has distinct epidemiologic or biological characteristics and a distinct genome that represents an independent replicating lineage”.102 Several circumstances have arisen that might seem to challenge these definitions, but as illustrated here, the definition allows resolution of the issues.
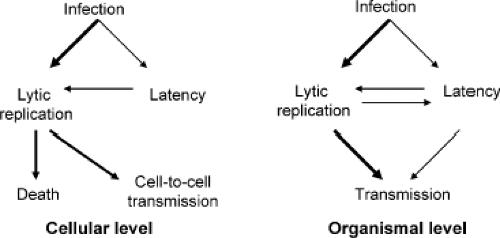 Figure 59.3. Outcomes of lytic and latent infections at the cellular and organismal levels. Thicker arrows represent more common events. |
The EBV variants, EBV-1 and EBV-2, differ markedly in several EBNA gene sequences. The differences lead to differences in some biological properties, including transforming potential. However, the variants do not occupy distinct ecological niches, the differences map to a small number of genes, and intermediates carrying one variant allele at one locus and the other variant allele at another locus have been detected. Thus, the EBV variants are recognized as allelic variants of the same species. At least two alleles have been identified of the KSHV K15 gene, which maps at the right end of the unique segment of the genome.105 As for the EBV variants, recombinants have been detected between viruses encoding the different K15 alleles; thus, this represents another example of intraspecies allelic variation. It is possible that the EBV and KSHV allelic variants represent early steps in speciation. The process of adaptation to a specialized niche would likely result in the gradual emergence of a new virus lineage characterized by additional mutations across its genome, a unique epidemiology, and reduced biological fitness of recombinants with the parental lineage. The net product would be a new virus species.
Species Nomenclature
Two forms of nomenclature are employed for herpesviruses: an informal (also known as vernacular or colloquial) nomenclature that often traces to the early days of virology, and a formal nomenclature that is sanctioned by the ICTV. For example, the virus informally known as Epstein-Barr virus (EBV) is formally known as Human herpesvirus 4. Table 59.1 and e-Table 59.2 include formal and informal names for each virus. In the ICTV-endorsed formal nomenclature, herpesvirus names consist of the family, subfamily, or genus of the natural host of the virus, the word “herpesvirus,” and a serial Arabic number (e.g., Cercopithecine herpesvirus 1; Table 59.1). Human herpesviruses are an exception to the host name rule (e.g., Human herpesvirus 7). ICTV-recognized virus species names are italicized, and the first letter of the first word of the name is capitalized.124 It has been proposed to rename herpesvirus species according to their subfamily, for example, Human herpesvirus 1 would be renamed as Human alphaherpesvirus 1, but this has not been formally approved.
The host name–serial number nomenclature system was adopted in 1973106a in an attempt to rectify problems associated with earlier systems in which viruses were named based on their disease associations, discoverer, geographic source, or whatever inspired the discoverer. Some viruses were given multiple names, and some names were applied to multiple viruses. Naming viruses after their associated diseases caused problems because
some viruses do not cause a specific disease, some viruses cause multiple and quite different diseases, and other viruses cause diseases whose etiologies are comprised of multiple agents. The formal nomenclature scheme was not meant to supplant the informal names that have been grandfathered by time and usage. The intent was to create an orderly system in which each virus would be named unambiguously and independently of classification or properties, which at that time were largely unknown. An important, and sometimes misunderstood, extension of this is that the species number is not intended to imply anything about the relationship between a virus and other herpesviruses that infect the same host species (e.g., HHV-7 and HHV-8 are members of different subfamilies) or between similarly numbered viruses that infect different host species (e.g., EHV-2 and BoHV-2 are members of different subfamilies).
some viruses do not cause a specific disease, some viruses cause multiple and quite different diseases, and other viruses cause diseases whose etiologies are comprised of multiple agents. The formal nomenclature scheme was not meant to supplant the informal names that have been grandfathered by time and usage. The intent was to create an orderly system in which each virus would be named unambiguously and independently of classification or properties, which at that time were largely unknown. An important, and sometimes misunderstood, extension of this is that the species number is not intended to imply anything about the relationship between a virus and other herpesviruses that infect the same host species (e.g., HHV-7 and HHV-8 are members of different subfamilies) or between similarly numbered viruses that infect different host species (e.g., EHV-2 and BoHV-2 are members of different subfamilies).
Table 59.1 Herpesviruses of Humans and of Veterinary or Scientific Importancea | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Classification
In the late 1970s, before viral DNA and amino acid sequences were known, herpesviruses were initially classified into one family (the Herpesviridae) and three subfamilies (the Alphaherpesvirinae, the Betaherpesvirinae, and the Gammaherpesvirinae) on the basis of biological properties.107 Remarkably, this framework remains useful and is supported by a deep and growing body of information extracted from nucleotide sequences. The viruses have been further classified into genera based on DNA sequence similarity, similarities in genome sequence
arrangement, and immunologic relatedness of important viral proteins.
arrangement, and immunologic relatedness of important viral proteins.
There has been an explosion of herpesviruses discovered through the use of degenerate polymerase chain reaction (PCR) primers that target highly conserved regions in core genes, such as the DNA polymerase.110 Most of this work has been done by assaying blood specimens, thus the list of known lymphotropic gammaherpesviruses has expanded greatly and additional betaherpesviruses have been identified. Expansion of the hunt to somewhat more difficult to obtain materials such as saliva or throat swabs and ganglia will likely yield a new array of novel viruses, including alphaherpesviruses. These new observations are important in that they thus far affirm the long-held hypothesis that individual animal species, or groups of closely related species, are hosts to particular herpesvirus species, with mammalian hosts being inhabited by viruses that represent each of the major mammalian herpesvirus lineages. Although the PCR amplimers sequenced in many of these studies are less than 400 base pairs (bp) long, the information is usually sufficient for reliable assignment of the novel virus to an established subfamily; genus-level assignments are less reliable in the absence of more extensive information.
Alphaherpesvirinae
This subfamily is defined on the basis of a variable host range, relatively short reproductive cycle, rapid spread in culture, efficient destruction of infected cells, and capacity to establish latent infections primarily—but not exclusively—in sensory ganglia. This subfamily contains the genera Mardivirus (GaHV-2), Iltovirus (GaHV-1), Simplexvirus (HSV-1), and Varicellovirus (VZV). Simplexviruses and Varicelloviruses have mammalian hosts, while Mardiviruses and Iltoviruses have avian hosts. Reptilian herpesviruses belong to the alphaherpesvirus lineage, but do not belong to any of the currently designated genera.76
Betaherpesvirinae
A nonexclusive characteristic of the members of this subfamily is a restricted host range. The reproductive cycle can be long (over 7 days), and the infection progresses slowly in cultured cells. Infected cells frequently become enlarged (cytomegalia), and carrier cultures are readily established. Betaherpesviruses can establish latency in secretory glands, lymphoreticular cells, kidneys, and other tissues. This subfamily contains the genera Cytomegalovirus (HCMV), Muromegalovirus (MCMV), Proboscivirus (ElHV-1), and Roseolovirus (HHV-6).
Gammaherpesvirinae
The host range of the members of this subfamily is restricted to the family or order of the natural host. In vitro all members replicate in lymphoblastoid cells, and some can lytically infect particular types of epithelioid and fibroblastic cells. Viruses in this group are usually specific for either T or B lymphocytes. Latency is ordinarily established in lymphoid tissue. This subfamily currently contains four genera: Lymphocryptovirus (EBV), Macavirus (AlHV-1), Percavirus (EHV-2), and Rhadinovirus (KSHV). Rhadinoviruses are mainly hosted by primates, Macaviruses are related to the ma lignant ca tarrhal fever viruses of ruminants, and the identified Percaviruses are hosted by per issodactyl and ca rnivore species. The lymphocryptoviruses consist of two major lineages that appear to have co-evolved with their hosts: viruses of Old World (humans, chimpanzees) and New World (marmosets) primates.43,77
Virion Architecture
The Virion
Mature herpesvirus virions vary in size from 120 to as much as 260 nm (reviewed in 109). The variation is in part due to variability in the thickness of tegument. Another major source of observed variability is the state of the envelope. Intact envelopes are impermeable and generally retain the quasispherical shape of the virion during preparation for electron microscopy (Fig. 59.1). Damaged envelopes are permeable to negative stains and the virions lose their quasispherical shape; permeated virions spread out on solid surfaces, resulting in a sunny-side-up egg appearance with a diameter much larger than for intact virions. The precise number of protein species contained in the virions is not known and may vary from one virus to another. Estimates based on analysis of purified virions in protein gels have generally been in the range of 35 to 45 major species. Proteomic analyses have identified from 24 to 71 virally encoded proteins in virions (4–7 in the nucleocapsid, 9–>20 in the tegument, and 4–19 in the envelope, plus a number of proteins whose location within virions is unknown).56,59,82,98,126 These and other studies also identified a number of host proteins in virion preparations, including moderately abundant quantities of cellular structural proteins, enzymes, and chaperones; actins and annexins have been consistently identified. The roles and necessity of the host-derived proteins in infection are not known. The abundance of individual protein species varies widely, from less than one copy per virion to over 1,000. Individual virions harbor on the order of 10,000 individual protein molecules. In addition to proteins, virions can include viral- and cellular-encoded messenger RNAs (mRNAs) that can be translated immediately after infection.11,114
While a lot is known about the average composition of populations of virions, the molecular definition of what constitutes an infectious virion remains to be defined. For example, gB is present in virion preparations at levels that correspond to an average of approximately 800 copies per virion. It is not known how widely the number of copies per virion ranges, or how many copies of gB are required to be present on a virion for it to be infectious. Importantly, most of the virus particles released from infected cells are not competent for virus replication but are bioactive in various ways. These include particles that appear by electron microscopy to be intact and complete virions of the sort just described. In addition, herpesviruses can produce large numbers of nonvirion particles of unknown biological significance, such as the dense bodies of HCMV, which are capsid-free, enveloped collections of tegument proteins.
Virion Components
The Core
The core of the mature virion contains a single molecule of the viral genome, in the form of nonchromatinized dsDNA that is packed in an orderly manner in the form of a torus.40,41,64,93,138 In some herpesvirions, the torus appears to be suspended by a proteinaceous spindle consisting of fibrils embedded in the underside of the capsid and passing through the hole of the torus. The precise arrangement of the DNA in the torus is not known, but the DNA is packed tightly, such that the internal volume of the capsid is approximately equal to the cylindrical volume of the genome. Because of the repulsive forces generated by the negatively charged phosphates that make up the backbone of the genome, such compact packing necessitates the abundant presence in the core of the anion spermine.45 Packaging of the viral genome into the capsid core requires ATP and results in a pressurized system that appears to be important for injection of virus genomes through the nuclear pore complex into nuclei of newly infected cells.63
The Capsid
The structural features of the capsid—that is, its 100-nm diameter, 161 capsomeres (150 hexons and 11 pentons), portal complex, and capsid triangulation number (T = 16)— are characteristic of all herpesviruses, including the distantly related fish and oyster viruses.9,31,64 Nonenveloped capsids are present in infected cells in three main forms: A-, B-, and C-capsids.44 A-capsids have no core structure, B-capsids contain the assembly scaffold but no genome, and C-capsids are DNA-containing species that no longer house the scaffold. The four conserved capsid proteins that comprise the major structural features of the capsid include the major capsid protein (MCP), the monomer and dimer proteins of the triplex (TRI1 and TRI2, respectively), and the small capsomere-interacting protein (SCP; HSV homologs are listed in Table 59.2). MCP is present in six copies per hexon (6 × 150 = 900 copies per capsid) and five per penton (5 × 11 = 55 copies per capsid), for a total of 955 copies per capsid. The triplex proteins interact with α2β stoichiometry and form complexes that are present at the 320 sites of threefold symmetry. Hexameric capsomeres are 9.5 × 12.5 nm in longitudinal section; a channel of 4 nm in diameter runs from the surface along their long axis.133 Penton channels are generally somewhat narrower and are nearly closed at their midpoint in B-capsids.135 The twelfth pentonal position is the portal for transit of genomic
DNA into and out of the capsid; it is composed of 12 copies of the capsid portal protein (PORT). The portal capping protein (PCP) is associated with mature, DNA-containing nucleocapsids. Cryoelectron microscopic image analysis has enabled reconstruction of capsid structures to greater than 10 Å resolution.139 Among other things, this has revealed a unique protein fold in the herpesvirus major capsid protein that is shared with the capsid proteins of tailed DNA bacteriophages, in support of a primordial linkage between these viruses.7 Beautiful animated representations of herpesvirus capsids and capsid components are available at http://www.eicn.ucla.edu/animations.
DNA into and out of the capsid; it is composed of 12 copies of the capsid portal protein (PORT). The portal capping protein (PCP) is associated with mature, DNA-containing nucleocapsids. Cryoelectron microscopic image analysis has enabled reconstruction of capsid structures to greater than 10 Å resolution.139 Among other things, this has revealed a unique protein fold in the herpesvirus major capsid protein that is shared with the capsid proteins of tailed DNA bacteriophages, in support of a primordial linkage between these viruses.7 Beautiful animated representations of herpesvirus capsids and capsid components are available at http://www.eicn.ucla.edu/animations.
The Tegument
The tegument, a term introduced by Roizman and Furlong109 to describe the proteinaceous structure between the nucleocapsid and the envelope, has no distinctive features in thin sections but may appear to be fibrous on negative staining.90,91,133 The tegument is sometimes distributed asymmetrically, and its thickness may vary, depending on the location of the virion within the infected cell. When the amount is variable, there is more of it in virions accumulating in cytoplasmic vacuoles than in those accumulating in the perinuclear space.40 Some evidence suggests that the amount of tegument is more determined by the virus than by the host.71 Teguments can contain more than 20 different virally encoded proteins, some of which are present at hundreds of copies per virion. Structural polarity across the tegument has been visualized by immunoelectron microscopy,120 indicating that it is an ordered structure, an observation supported by cryoelectron microscopic observations of tegument–nucleocapsid interactions.123,134 This is further evidenced by tegument proteins that are closely associated with the nucleocapsid (inner tegument) being acquired in the nucleus and by interactions between envelope glycoproteins and tegument proteins at the tegument periphery. Following nuclear egress, subsequent components of the tegument are likely added in a somewhat ordered manner as the virus particle matures during its trek through the cytoplasm.80 One purpose of the tegument is to carry into newly infected cells an assortment, or toolbox, of already synthesized proteins that can immediately begin to manage the host environment to meet the needs of the virus, such as by shutting down host protein synthesis, inhibiting infection-triggered cell defenses, and stimulating viral gene expression.
The Envelope
Electron microscopic studies on thin sections have shown that the outer covering of the virion, the envelope, has a typical trilaminar appearance.38 The presence of lipids was demonstrated by analyses of virions5 and by the sensitivity of the virions to lipid solvents and detergents.118 Virion envelopes are derived from patches of altered membrane that trace to the organelle where envelopment occurs.3,40,66,91 A major constituent of virion envelopes is a collection of virally encoded glycoproteins. The number and relative amounts of viral glycoproteins vary among herpesviruses. HSV specifies at least 11 different virion-associated glycoproteins, and the copy number of individual glycoproteins can exceed 1,000 per virion. The glycoproteins form numerous protrusions on virion envelopes that are more numerous and shorter than those present on the surface of many other enveloped viruses.133
Genomic and Genetic Architecture
Genome Size and Base Composition
Herpesvirus DNAs extracted from virions and characterized to date are linear and double stranded, but they circularize immediately on release from nucleocapsids into the nuclei of infected cells.
Distinguishing features of herpesvirus DNAs include their length and base composition. The length of herpesvirus DNAs varies from approximately 124 to 295 kbp (Table 59.1 and e-Table 59.2). The variability in genome lengths of different herpesviruses is distinct from the generally less extensive polymorphism in the size of DNAs of individual viruses. Thus, herpesvirus genomes contain terminal and internal reiterated sequences that can vary in copy number, as well as sequences that can be lost or duplicated during passage in cell culture, leading to intraspecies variation in genome lengths that can exceed 10 kbp.
The base composition of herpesvirus DNAs varies from 31% to 77% total G+C content Table 59.1 and e-Table 59.2). Furthermore, herpesvirus DNAs vary with respect to the extent of homogeneity of G+C content across the length of the genome (generally higher G+C composition in terminal repeats). The extent of inhomogeneity in the base composition varies from minimal (e.g., HSV) to very extensive. For example, the genome of MuHV-4 has a G+C content of 46% in its unique region and 78% in its terminal repeats.130
The genetic requirements for efficient replication in cultured cells can differ from the in vivo requirements. Spontaneous deletions have been noted in HSV and EBV strains passaged outside the human host (e.g., EBV strain P3HR1, HSV-1 strain HFEM). Highly passaged strains of HCMV lack a segment encoding at least 19 genes that are present in wild-type isolates.17 A progression of changes in a small number of genes enables clinical isolates of HCMV to begin replicating efficiently in cultured cells.23
Stay updated, free articles. Join our Telegram channel

Full access? Get Clinical Tree