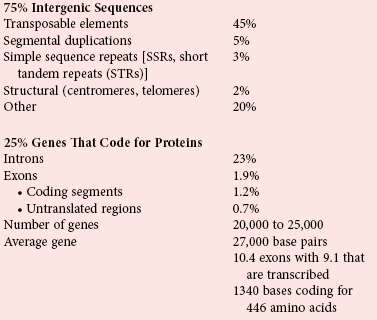
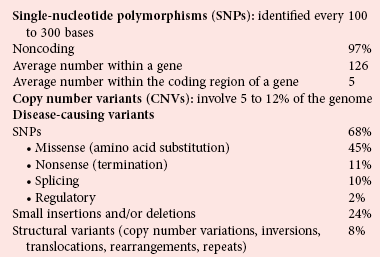
Chapter 38 Each human cell contains two copies of a 3 billion member sequence code of nucleic acids on 46 chromosomes.4,5,12 Box 38-1 lists statistics for the human genome and the types of variations that are important in clinical diagnostics. Although intergenic DNA does not code for protein and was originally considered “junk,” much of this DNA is transcribed to RNA, producing a complex “transcriptome” network of RNA control elements whose function and mechanics are active areas of investigation.1 Even though 99% of the genome does not code for protein, most of it is transcribed into noncoding RNA. At least 93% of the genome is transcribed,1 producing more than 10 times the amount of RNA that is produced from the coding segments of genes.2 Both strands of DNA may be transcribed, and long noncoding transcripts may overlap coding regions, producing a complex transcriptome of functional RNA molecules that may variably regulate transcription of coding regions, RNA processing, mRNA stability, translation, protein stability, and secretion. In addition to long noncoding RNA, ribosomal RNA, and transfer RNA, specific classes of noncoding RNAs include small nuclear RNAs critical for splicing, small nucleolar RNAs that modify rRNA, telomerase RNAs for maintenance of telomeres, small interfering RNAs, and microRNAs that regulate gene expression.8,9 MicroRNAs (or miRNAs) are noncoding but functional single-stranded RNAs that are about 22 bases long and are expressed in a tissue-specific manner. They are initially transcribed as longer precursors that undergo two rounds of truncations as they are transported from nucleus to cytoplasm in the cell. The mature miRNA is then integrated into a protein complex called the RNA-induced silencing complex, which regulates translation of mRNA. MicroRNAs hybridize to a 6 to 8 base sequence in the 3′ untranslated region of a target mRNA and inhibit mRNA expression, by mRNA degradation if the remaining bases are perfectly complementary, or by blocking of translation if they are imperfectly complementary. More than 700 different miRNAs have been reported,3 and sequences encoding for miRNA have been found on every chromosome except the Y chromosome. Consider the genome as a book. Nucleotides are the individual letters, and three bases make up each word as an amino acid codon. The words are organized into sentences or exons that are separated by periods or introns. Each sentence is further organized into paragraphs or genes. Many paragraphs constitute a chapter or chromosome, and several chapters make up a book or genome. If the DNA of any two individuals is compared, on average one spelling difference is noted every 1250 bases (i.e., approximately 99.9% of the sequence is identical between randomly chosen copies of the genome). However, different individuals (copies of the same book) vary in a subtler way. Some of the pages are copied more than once and may be scattered throughout the book. Such copy number variants involve a greater amount of text than the spelling differences, with 0.5% of the genome differing on average between two individuals when 50 kb pages are considered,7 that is, between individuals, at least five times as many bases are affected by copy number changes than by small sequence differences. Although SNPs are the most common sequence variant, copy number variants cover more of the genome than SNPs. These copy number variants (CNVs) occur in stretches of DNA that may range from 100 bases up to several Mb (megabases, or million bases) in size. CNVs may be duplicated in tandem or may involve complex gains or losses of homologous sequences at multiple sites in the genome. CNV regions exist in every chromosome and involve 5 to 12% of the human genome.7,10 Most CNVs are inherited and biallelic, similar to SNPs.7 More than 6000 CNV loci have been reported, and many of them overlap with genes. Individuals differ on average at more than 200 CNV loci, and these overlap the transcribed regions of more than 100 genes.7
Genomes and Nucleic Acid Alterations
Human Genome
Variation Within the Human Genome
![]()
Stay updated, free articles. Join our Telegram channel

Full access? Get Clinical Tree


Basicmedical Key
Fastest Basicmedical Insight Engine