Genome Structure, Regulation, and Tissue Differentiation
Jacquelyn L. Banasik
Key Questions
• How is genetic information stored in the cell and transmitted to progeny during replication?
• What roles do genes play in determining cell structure and function?
• How is gene expression regulated?
![]()
http://evolve.elsevier.com/Copstead/
The ability of scientists to study and manipulate genes has evolved at an incredible pace, including the complete sequencing of all 6.4 billion nucleotides in an entire human genome. A better understanding of the role that genetics plays in cellular function and disease has spurred efforts to develop therapies to correct genetic abnormalities. The science of genetics developed from the premise that invisible, information-containing elements called genes exist in cells and are passed on to daughter cells when a cell divides. The nature of these elements was at first difficult to imagine: what kind of molecule could direct the daily activities of the organism and be capable of nearly limitless replication? The answer to this question was discovered in the late 1940s and was almost unbelievable in its simplicity. It is now common knowledge that genetic information is stored in long chains of stable molecules called deoxyribonucleic acid (DNA). The human genome contains approximately 23,000 genes encoded by only four different molecules. These molecules are the deoxyribonucleotides containing the bases adenine (A), cytosine (C), guanine (G), and thymine (T). Genes are composed of varying sequences of these four bases, which are linked together by sugar-phosphate bonds. By serving as the templates for the production of body proteins, genes ultimately affect all aspects of an organism’s structure and function. When the sequencing of an entire human genome was completed in 2004 it became clear that the genome is much more complex than the sum of its genes. Only 1.3% of chromosomal DNA codes for proteins and many DNA sequences code for ribonucleic acid (RNA) molecules that function in the nucleus to regulate gene function. Methods to rapidly survey the DNA sequences of a particular person are available and genetics is an increasingly important consideration in the etiology, pathogenesis, and pharmacologic treatment of a variety of diseases. However, genetic inheritance involves more than the transfer of genes from parent to offspring. For example, the nutritional exposures of grandparents may influence the metabolic physiology of grandchildren through a process known as epigenetics. Epigenetics is further explored in Chapter 6. Knowledge of the basic principles of genetics and gene regulation is a prerequisite to understanding not only conventional genetic diseases but also nearly every pathophysiologic process. This chapter examines the biochemistry of genes (molecular genetics), the regulation of gene expression, and the processes of tissue differentiation. Principles of genetic inheritance precede the discussion of genetic diseases in Chapter 6.
Molecular Genetics
Structure of DNA
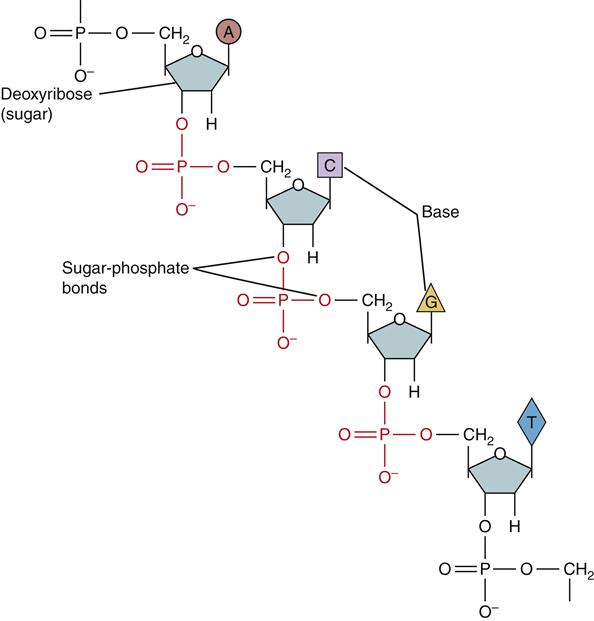
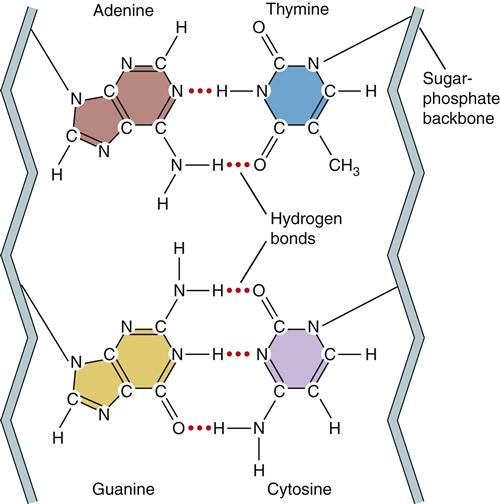
In humans, DNA encodes genetic information in 46 long double-stranded chains of nucleotides called chromosomes.1 The nucleotides consist of a 5-carbon sugar (deoxyribose), a phosphate group, and one of the four nucleotide bases (Figure 5-1). The nucleotide bases are divided into two types based on their chemical structure. The pyrimidines, cytosine and thymine, have single-ring structures. The purines, guanine and adenine, have double-ring structures (Figure 5-2). DNA polymers are formed by the chemical linkage of these nucleotides. The sugar-phosphate linkages, also called phosphodiester bonds, join the phosphate group on one sugar (attached to the 5-carbon) to the 3-carbon of the next sugar (see Figure 5-1). The four kinds of bases (A, C, G, T) are attached to the repeating sugar-phosphate chain. The bases of one strand of DNA form weak bonds with the bases of another strand of DNA. These noncovalent hydrogen bonds are specific and complementary (Figure 5-3). The bases G and C always bond together and the bases A and T always bond together. Nucleotides that are able to bond together in this complementary way are called base pairs.
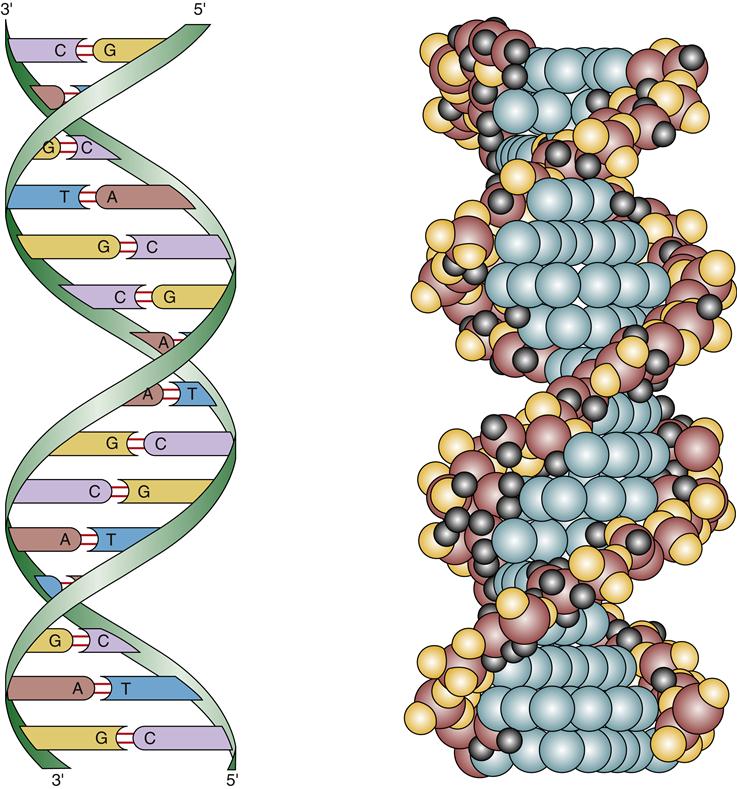
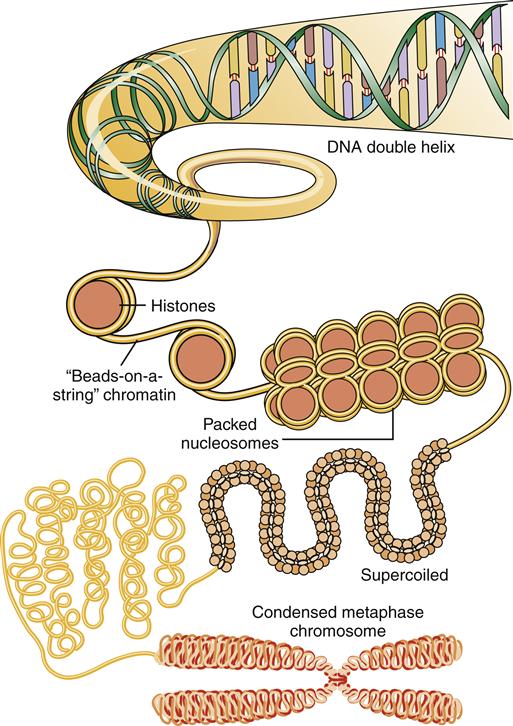
In the early 1950s, Watson and Crick proposed that the structure of DNA was a double helix.2 In this model, DNA can be envisioned as a twisted ladder, with the sugar-phosphate bonds as the sides of the ladder and the bases forming the rungs (see Figure 5-3). There is one complete turn of the helix every 10 base pairs. The two strands of DNA must be complementary to form the double helix; that is, the bases of one strand must pair exactly with their complementary bases on the other strand. The helix is wound around proteins called histones to form nucleosomes (Figure 5-4). DNA coupled to histones and other nuclear proteins is termed chromatin. When a cell is not dividing, the chromatin is loosely packed within the nucleus and not visible under the light microscope. During cell division, the chromatin becomes tightly condensed into the 46 chromosomes that become visible during mitosis.
The discovery of the double-helix model was profound because it immediately suggested how information transfer could be accomplished by such simple molecules. Because each DNA strand carries a nucleotide sequence that is exactly complementary to the sequence of its partner, both strands can be used as templates to create an exact copy of the original DNA double helix. When a cell divides to form two daughter cells, each daughter cell must receive a complete copy of the parent cell’s DNA. The process of DNA replication requires separation of the DNA double helix by breaking the hydrogen bonds between the base pairs. Specific replication enzymes then direct the attachment of the correct (complementary) nucleotides to each of the single-stranded DNA templates. In this way, two identical copies of the original DNA double helix are formed and passed on to the two daughter cells during cell division.
DNA Replication
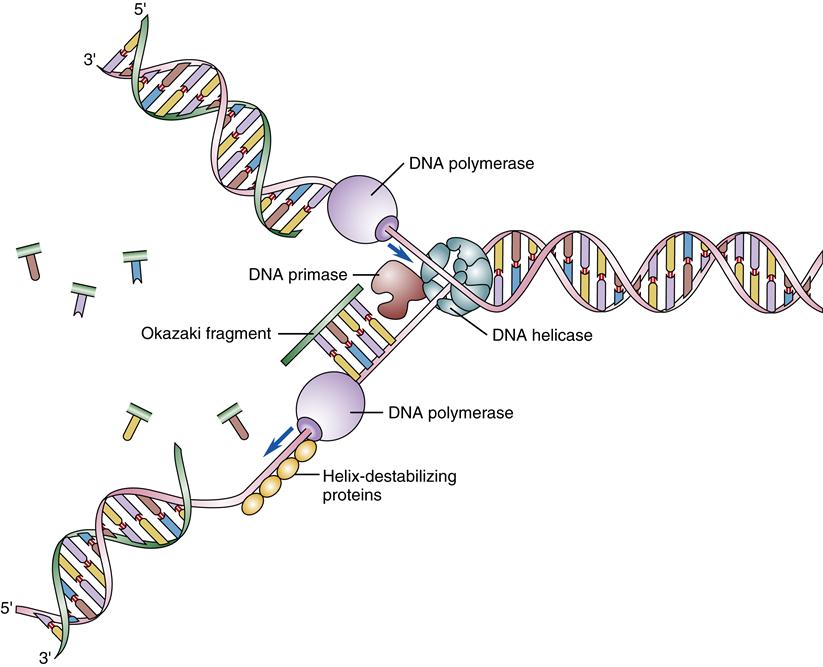
Although the underlying principle of gene replication is simple, the cellular machinery required to carry out the replication process is complex, involving a host of enzymes and proteins.3 These “replication machines” can duplicate DNA at a rate of 1000 nucleotides per second and complete the duplication of the entire genome in about 8 hours.4 The DNA double helix must first separate so that new nucleotides can be paired with the old DNA template strands. The DNA double helix is normally very stable: the base pairs are locked in place so tightly that they can withstand temperatures approaching the boiling point. In addition, DNA is wrapped around histones and bound by a host of DNA-binding proteins through which the replication machinery must navigate. DNA replication is started by special proteins (initiator proteins) that pry the DNA strands apart at specific places along the chromatin, called replication origins. Then special enzymes (DNA helicases) are needed to rapidly unwind and separate the DNA strands, whereas helix-destabilizing proteins (also called single-stranded DNA-binding proteins) bind to the exposed DNA strands to keep them apart until replication can be accomplished (Figure 5-5). As the DNA is unwound in the replication fork, it becomes overly twisted downstream, so another set of enzymes, topoisomerases, cuts nicks in the DNA and allows it to unwind to prevent tangling. Ligases repair the nicks.
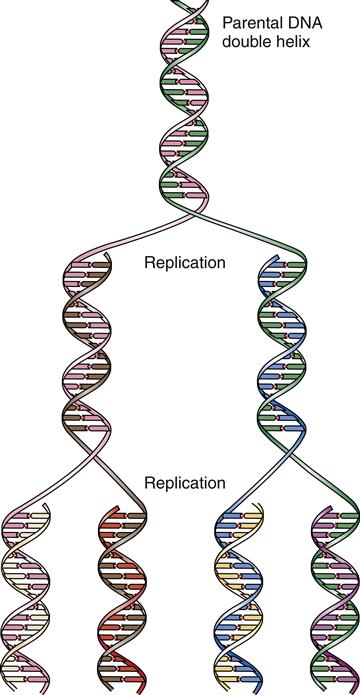
Once a portion of the DNA double helix has been separated, an enzyme complex, DNA polymerase, binds the single strands of DNA and begins the process of forming a new complementary strand of DNA. The polymerases match the appropriate base to the template base and catalyze the formation of the sugar-phosphate bonds that form the backbone of the DNA strand. Replication proceeds along the DNA strand in one direction only: from the 3′ end toward the 5′ end.4 The ends of the DNA strands are labeled 3′ and 5′ according to the exposed carbon atom at that end. Because two complementary DNA strands are antiparallel, DNA replication is asymmetrical; one strand, the leading strand, is replicated as a continuous polymer, but the lagging strand must be synthesized in short sections in a “backstitching” process (see Figure 5-5). The backstitched fragments of DNA, called Okazaki fragments, are then sealed together by DNA ligase to form the unbroken DNA strand. DNA polymerase is unable to replicate DNA located at the very ends of the chromosomes (the telomeres), so another special enzyme complex, telomerase, is needed for this. The telomeres are fairly short, being composed of approximately 1000 repeats of a GGGTTA sequence. When the telomeres are replicated, one side of the double helix (3′ end) is always longer and loops around and tucks back into the strand. This prevents nuclear enzymes from mistaking the ends of the chromosomes as broken DNA ends and trying to attach them to each other. In many somatic cell types, telomerase activity is low and the cell’s chromosomes become slightly shorter with each cell division. Chromosomal shortening has been proposed as a mechanism of “counting” the number of replications and may be important in cellular aging and prevention of cancer (see Chapter 7). DNA replication is said to be semiconservative because each of the two resulting DNA double helices contains one newly synthesized strand and one original (conserved) strand (Figure 5-6).
The DNA polymerase also has the ability to proofread the newly synthesized strands for errors in base pairing. If an error is detected, the enzyme will reverse, remove the incorrect nucleotide, and replace it with the correct one. The fidelity of copying during DNA replication is such that only about one error is made for every 109 base pair replications.5 The self-correcting function of the DNA polymerases is extremely important because errors in replication will be transmitted to the next generation of cells.
Genetic Code
How do an organism’s genes influence its structural and functional characteristics? A central theory in biology maintains that a gene directs the synthesis of a protein. It is the presence (or absence) and relative activity of various structural proteins and enzymes that produce the characteristics of the cell. This definition of genes as protein-coding elements is not entirely correct because many “genes” code for ribonucleic acid (RNA) molecules as their final functional products and some genes may code for more than one protein product through alternate splicing of the RNA messages. Protein synthesis still holds a predominant place in understanding how genes direct cell structure and function. One of the surprising outcomes of the Human Genome Project was how little of the DNA in chromosomes contains coding segments (less than 2%) and the low number of genes that exist (23,000). Before the completion of the Human Genome Project, it was estimated that the human genome contained 100,000 genes.
Proteins are composed of one or more chains of amino acids (polypeptides) that fold into complex three-dimensional structures. Cells contain 20 different types of amino acids that connect in a specific sequence to form a particular protein (Table 5-1). Each type of protein has a unique sequence of amino acids that dictates its structure and activity.
TABLE 5-1
RNA CODONS FOR THE DIFFERENT AMINO ACIDS AND FOR START AND STOP
| AMINO ACIDS | RNA CODONS | |||||
| Alanine | GCU | GCC | GCA | GCG | ||
| Arginine | CGU | CGC | CGA | CGG | AGA | AGG |
| Asparagine | AAU | AAC | ||||
| Aspartic acid | GAU | GAC | ||||
| Cysteine | UGU | UGC | ||||
| Glutamic acid | GAA | GAG | ||||
| Glutamine | CAA | CAG | ||||
| Glycine | GGU | GGC | GGA | GGG | ||
| Histidine | CAU | CAC | ||||
| Isoleucine | AUU | AUC | AUA | |||
| Leucine | CUU | CUC | CUA | CUG | UUA | UUG |
| Lysine | AAA | AAG | ||||
| Methionine | AUG | |||||
| Phenylalanine | UUU | UUC | ||||
| Proline | CCU | CCC | CCA | CCG | ||
| Serine | UCU | UCC | UCA | UCG | AGC | AGU |
| Threonine | ACU | ACC | ACA | ACG | ||
| Tryptophan | UGG | |||||
| Tyrosine | UAU | UAC | ||||
| Valine | GUU | GUC | GUA | GUG | ||
| Start (CI) | AUG | |||||
| Stop (CT) | UAA | UAG | UGA | |||
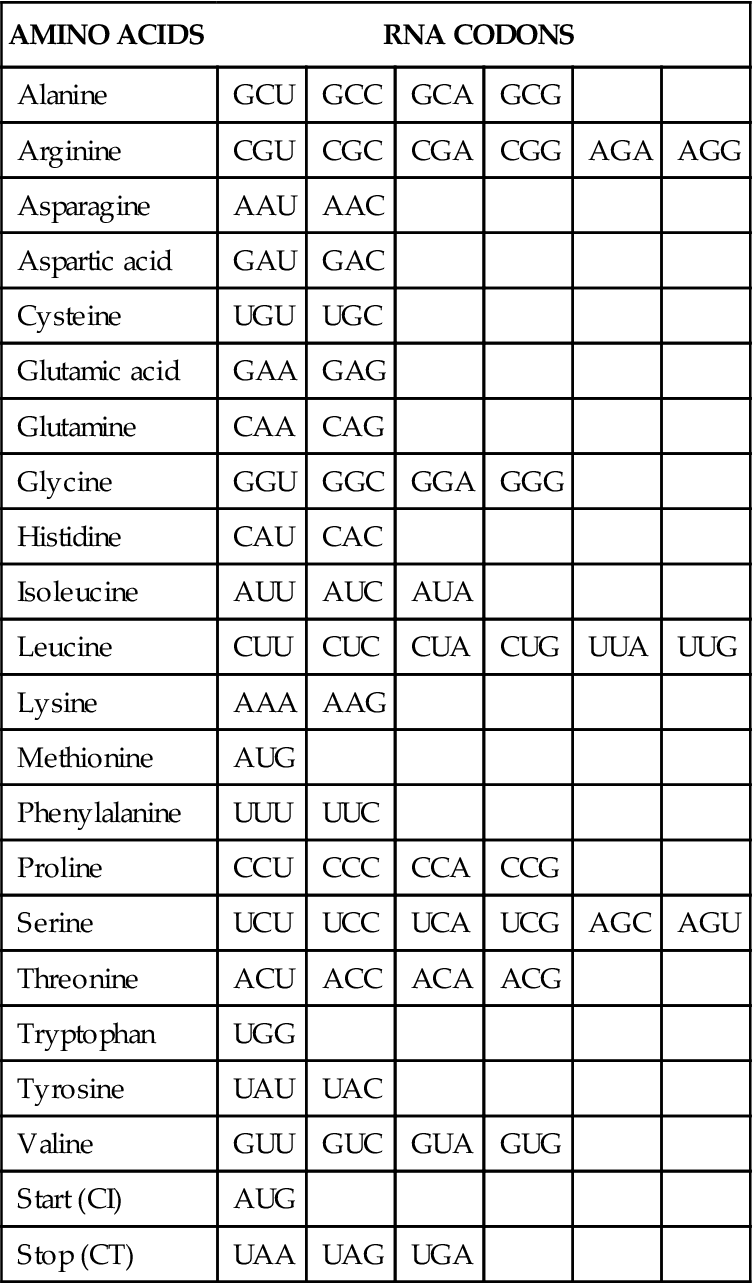
If genes are to direct the synthesis of proteins, the information contained in just four kinds of DNA nucleotide bases must code for 20 different amino acids. This so-called genetic code was deciphered in the early 1960s.6,7 It was determined that a series of three nucleotides (triplet) was needed to code for each of the 20 amino acids. Because there are four different bases, there are 43, or 64, different possible triplet combinations. This is far more than needed to code for the 20 known amino acids. Three of the nucleotide triplets or codons do not code for amino acids and are called stop codons because they signal the end of a protein code. The remaining 61 codons code for 1 of the 20 amino acids (see Table 5-1). Obviously, some of the amino acids are specified by more than one codon. For example, the amino acid arginine is determined by six different codons. The code has been highly conserved during evolution and is essentially the same in organisms as diverse as humans and bacteria.
Several intermediate molecules are involved in the process of DNA-directed protein synthesis, including the complex protein-synthesizing machinery of the ribosomes and several types of RNA. RNA is structurally similar to DNA, except that the sugar molecule is ribose rather than deoxyribose, and one of the four bases is different in that uracil replaces thymine. Because of the biochemical similarity of uracil and thymine, both can form base pairs with adenine. In addition, RNA can form stable single-stranded molecules, whereas DNA strands anneal together, forming a double-stranded molecule.
Several functionally different types of RNA are involved in protein synthesis and cell function. The number and variety of RNA molecules existing within the nucleus is large (Box 5-1) and the exact function of most has yet to be determined. Some perform messenger RNA (mRNA) splicing, ribosome assembly, and quality control of RNA messages before they are transferred to the cytoplasm. The roles of three types of RNA that participate in protein production are well understood. Ribosomal RNA (rRNA) is found associated with the ribosome (see Chapter 3) in the cell cytoplasm. Messenger RNA is synthesized from the DNA template in a process termed transcription and carries the protein code to the cytoplasm, where the proteins are manufactured. The amino acids that will be united to form proteins are carried in the cytoplasm by a third type of RNA, transfer RNA (tRNA), which interacts with mRNA and the ribosome in a process termed translation.
Transcription
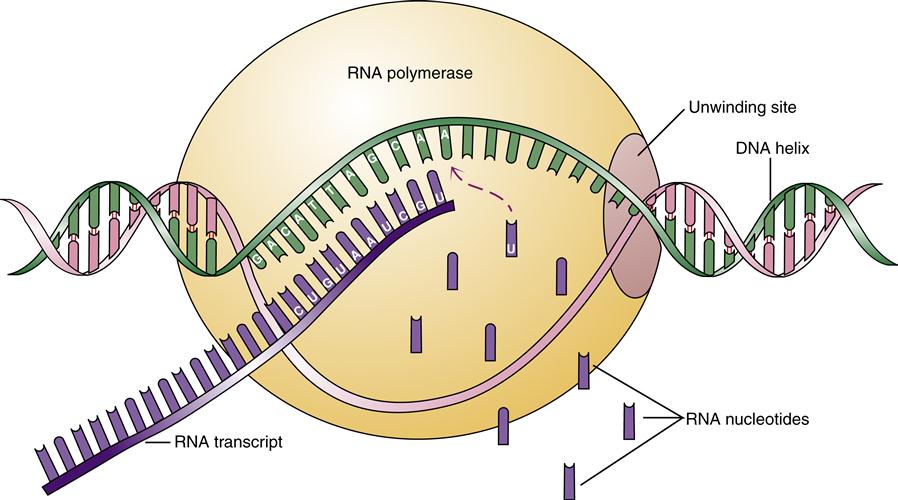
Transcription is the process whereby mRNA is synthesized from a single-stranded DNA template. The process is similar in some respects to DNA replication. Double-stranded DNA must be separated in the region of the gene to be copied, and specific enzyme complexes (DNA-dependent RNA polymerases) orchestrate the production of the mRNA polymer. Only one of the DNA strands contains the desired gene sequence and serves as the template for the synthesis of mRNA. This strand is called the sense strand. The other strand is termed the nonsense or antisense strand and is not transcribed into an RNA message.
Some genes are continuously active in certain cells, whereas others are carefully regulated in response to cellular needs and environmental signals. Special sequences of DNA near a desired gene may enhance or inhibit its rate of transcription. In general, a gene is transcribed when the RNA polymerase–enzyme complex binds to a promoter region just upstream of the gene’s start point. This binding event requires the cooperative function of numerous DNA-binding proteins. Once bound at the promoter, the RNA polymerase directs the separation of the DNA double helix and catalyzes the synthesis of the RNA message by matching the appropriate RNA bases to the DNA template (Figure 5-7). The RNA message is directly complementary to the DNA sequence, except that uracil replaces thymine.
Stay updated, free articles. Join our Telegram channel

Full access? Get Clinical Tree