CHAPTER 15 Gene Expression*
Each organism, whether it has 600 genes (Mycoplasma), 6000 genes (budding yeast), or 25,000 genes (humans), depends on reliable mechanisms to turn these genes on and off. This is called regulation of gene expression. In simple organisms, such as bacteria and yeast, environmental signals, such as temperature or nutrient levels, control much of gene expression. In multicellular organisms, genetically programmed gene expression controls development from a fertilized egg. Within these organisms, cells send each other signals that control gene expression either through direct contact or via secreted molecules, such as growth factors and hormones.
The Transcription Cycle
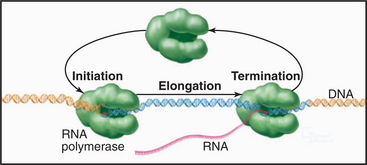
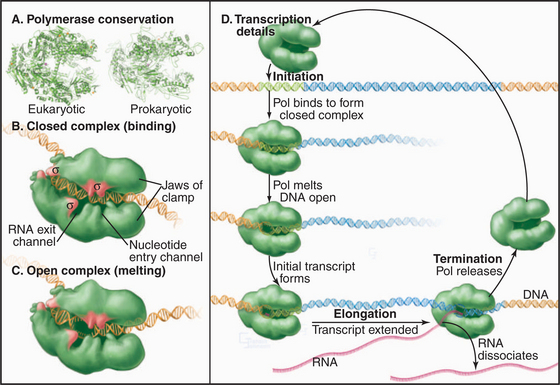
Synthesis of RNA by RNA polymerases is a cyclic process that can be broken down into three sets of events: initiation, elongation, and termination (Fig. 15-1). Each of these events consists of multiple individual steps. In the first step of the initiation process, RNA polymerase locates and binds to the chromosome near the beginning of the gene, forming a preinitiation complex at a sequence termed a promoter. This binding must be highly specific to distinguish promoter from nonpromoter DNA. Next, a conformational change in the polymerase-promoter complex results in formation of an open complex in which the DNA duplex is unpaired, allowing RNA polymerase access to nucleotide bases that are complementary to the start of the message. After formation of a phosphodiester bond between the first two complementary ribonucleotides, the polymerase translocates one base and repeats the process of phosphodiester bond formation, resulting in elongation of the nascent RNA. The elongation reaction cycle continues at an average rate of about 20 to 30 nucleotides per second until the complete gene has been transcribed. Elongation is not a uniform reaction, however, as RNA polymerase pauses at certain sequences. These pauses are important for regulation of transcription. The final step in the transcription cycle, termination, occurs when the polymerase reaches a signal on DNA that causes an extended pause in elongation. Given enough time and the appropriate sequence context, the nascent transcript dissociates from the elongating RNA polymerase, and the DNA template returns to a base-paired duplex conformation. Ultimately, RNA polymerase dissociates from the template and is free to begin a new search for a promoter.
Each of the steps in the transcription cycle can potentially serve as the target of regulatory molecules. The frequency of initiation varies among different promoters as dictated by the need for the gene product. The initiation reaction is most often regulated, presumably because this prevents synthesis of messages that encode unneeded products. Elongation and termination can also be regulated, as can splicing and further processing of mRNAs (see Chapter 16). In eukaryotes, the sum of these nuclear regulatory steps, together with cytoplasmic regulation of mRNA stability and translation efficiency, contributes to the wide variation seen in the abundance of different mRNAs and proteins in particular types of cells.
The Transcription Unit
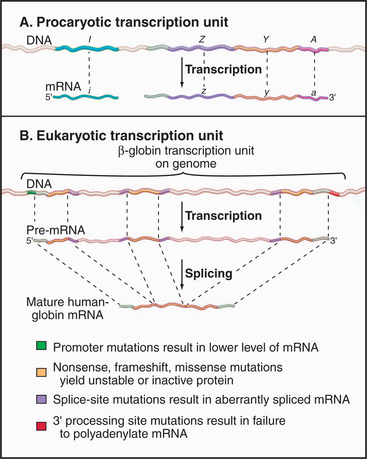
Coding information in genomes is transcribed in increments corresponding to one or a few genes. Gene-coding and regulatory (cis-acting) DNA sequences that direct transcription initiation, elongation, and termination are collectively called a transcription unit. Prokaryotic transcription units, called operons, contain more than one gene, often encoding physiologically related proteins (Fig. 15-2A). Operons are flanked by sequences that direct the initiation and termination of transcription. Figure 15-2B shows a simple eukaryotic transcription unit encoding the human hemoglobin β-chain. Although only a small fraction of this region encodes the β-globin polypeptide, the adjacent regulatory se-quences are crucial for proper expression of β-globin. Genetic defects resulting in decreased β-globin production are called β-thalassemias. Such mutations can occur either in the coding region, resulting in an unstable or truncated polypeptide, or in the adjacent control regions, leading to low levels of transcription or aberrant processing of the newly synthesized RNA (see Chapter 16). Thus, the transcription unit can be thought of as a linked series of modules, all of which must be functional for the gene to be transcribed at the correct level.
Biogenesis of RNA
Transcription of eukaryotic DNA in the nucleus is linked to subsequent steps that process the nascent transcript in preparation for its eventual function (see Chapter 16 for a complete discussion of these steps). For mRNA precursors, this includes capping and methylation of the 5′ end of the nascent transcript. Most messages are also spliced to remove introns; the 3′ end of the message is then cleaved, and a stretch of adenosine residues is added. The mRNA is then transported to the cytoplasm, where it serves as the template for protein synthesis.
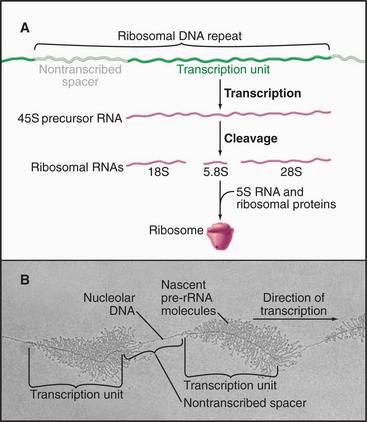
Eukaryotic ribosomal RNA is synthesized from a set of tandemly repeated genes as a single molecule, which is cleaved and modified to give the final 28S, 5.8S, and 18S RNAs (Fig. 15-3). These are assembled, together with 5S RNA and about 80 proteins, into ribosomes in the nucleolus. Transfer RNA is synthesized in the nucleus and transported to the cytoplasm, where it is charged with amino acids prior to participating in protein synthesis (see Chapter 17). snRNAs are synthesized and processed in the nucleus. From there, they migrate to the cytoplasm, where they acquire essential proteins, and then return to the nucleus, where they function in the enzymatic reactions of RNA processing (splicing; see Chapter 16). The postsynthetic processing pathway that a particular transcript follows is dictated, in part, by the transcription machinery that is used to initiate and elongate the transcript and by certain features of the nascent RNA.
RNA Polymerases
RNA polymerases synthesize a new strand of nucleic acid that is complementary to one of the chromosomal DNA strands. While the enzymatic reaction is similar to DNA replication (see Chapter 42), there are several important differences. First, RNA polymerases synthesize a strand of ribonucleotides. Second, unlike DNA polymerase, RNA polymerases can initiate transcription without a primer. Finally, unlike replication, the newly transcribed sequences do not remain base-paired with the template but are displaced after reaching a length of about 10 nucleotides. These properties are common to RNA polymerases in all cells; therefore, it is not surprising that all cellular RNA polymerases share common structural features.
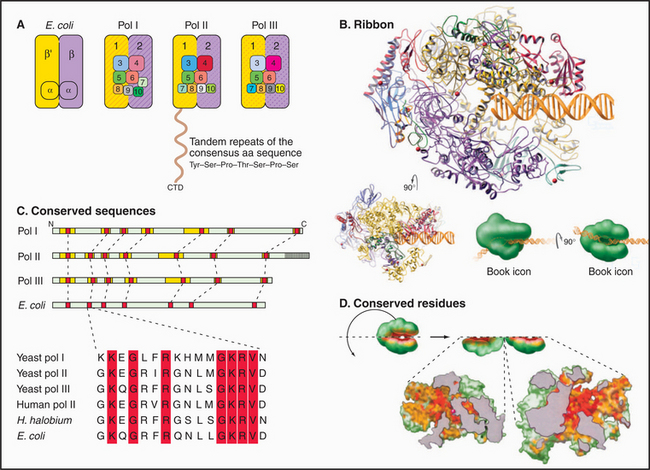
Most eukaryotes have three different RNA polymerases (some species of plants contain four). The largest subunits of the three eukaryotic RNA polymerases are closely related to the bacterial β and β′ subunits. RNA polymerases I, II, and III have up to 10 additional subunits, most of which are unique to each enzyme (Fig. 15-4A). The subunits of both prokaryotic and eukaryotic enzymes assemble into a structure that is roughly spherical, with a diameter of approximately 150 Å and a 25-Å-wide cleft, large enough to accommodate the DNA template (Fig. 15-4B). The site of nucleotide addition is located on the back wall of the cleft. The framework of this structure is provided by the two largest subunits, which make up the two lobes that clamp down on the template DNA.
The eukaryotic polymerases can be distinguished experimentally on the basis of their sensitivity to the fungal toxin α-amanitin, RNA polymerase II being the most sensitive and RNA polymerase I being the most resistant. RNA polymerase I localizes to the nucleolus, where it synthesizes rRNA. RNA polymerase II synthesizes mRNA and several snRNAs involved in RNA splicing in the nucleoplasm. RNA polymerase III synthesizes tRNA, 5S rRNA, and the 7S RNA of the signal recognition particle (see Fig. 20-5). The newly described RNA polymerase IV is present in plants, where it is involved in heterochromatin formation and gene silencing.
Specialization has been balanced, however, by the need to retain the structural elements required for RNA synthesis. In each eukaryotic RNA polymerase, the largest subunits are homologous to the bacterial β′ – and β-subunits that make up the catalytic core of prokaryotic RNA polymerases (Fig. 15-4C). The structure of a bacterial RNA polymerase reveals that the most conserved residues are located on the inner surfaces of the enzymes, where they are likely to be involved in the synthesis of RNA (Fig. 15-4D).
RNA Polymerase Promoters
Initiation of transcription requires RNA polymerase loading onto the chromosome at the promoter of a gene or operon. The promoter can be loosely defined as the sum of DNA sequences necessary for transcription initiation. This definition is not sufficient, however, as most genes are regulated (positively or negatively) at the transcription initiation level. In eukaryotic cells, packaging into chromatin represses most promoters, and activator proteins are required for recruiting RNA polymerase to the site of initiation. In prokaryotes, both activators and repressors modulate the frequency of initiation at promoters. Strong promoters drive the expression of genes whose products are required in abundance, whereas weaker promoters are selected for expression of rare proteins or RNAs. In multicellular organisms, a promoter may direct expression at an intermediate level in some cells, at an activated level in others, and at a repressed level in yet others.
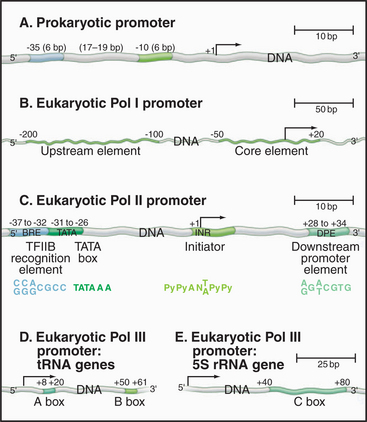
Promoters in bacteria are recognized by direct interactions between specific DNA sequences and the RNA polymerase σ factor. The most common σ factor in E. coli (σ 70) recognizes two conserved six-base sequences located 10 bases (minus 10) and 35 (minus 35) upstream of the transcription start site (Fig. 15-5A). Once initiation has occurred, σ is no longer required and can dissociate from the core enzyme. Bacterial cells have several distinct σ factors, each of which binds the core enzyme and direct RNA polymerase to a subset of promoters that contain different recognition sequences, thereby promoting transcription of genes with related functions.
Eukaryotic RNA polymerase I and II promoter sequences are also situated upstream of the transcription start site. In contrast, RNA polymerase III promoters contain key promoter elements within the transcribed sequences. RNA polymerase I recognizes a single type of promoter located upstream of each copy of the long tandem array of pre-rRNA coding sequences (Fig. 15-5B). The core element of this promoter overlaps the transcription start site, while an upstream control element located approximately 100 base pairs (bp) from the start site stimulates transcription. RNA polymerase I is not required in yeast cells that contain a pre-rRNA gene under control of an RNA polymerase II promoter. Therefore, if RNA polymerase I does recognize other promoters, these transcripts are not required for viability.
Comparison of the first eukaryotic protein-coding gene sequences revealed a conserved consensus se-quence located approximately 30 bp upstream of the transcription start site of many RNA polymerase II–transcribed genes (Fig. 15-5C). This consensus sequence—TATAAAA—called a TATA box, shows some similarity to the bacterial -10 sequence. In addition to the TATA box, a less conserved promoter element, the initiator, is found in the vicinity of the transcription start site of many genes. RNA polymerase II–transcribed genes that do not contain TATA boxes often contain strong initiator elements. Together, these two elements account for the basal promoter activity of most protein-coding genes.
Both types of RNA polymerase III promoters have key elements within the transcribed sequences (Fig. 15-5D–E). tRNA genes contain two 11-bp elements, the A box and B box, centered about 15 bp from the 5′ and 3′ ends of the coding sequence, respectively. The 5S-rRNA gene contains a single internal element, the C box, located in the center of the coding region. Given the differences in classes of eukaryotic promoters, it is not surprising that different polymerases use different proteins to recognize the promoter sequences.
Transcription Initiation
The loading of RNA polymerase onto the double-stranded genomic DNA at a promoter sequence is best understood in prokaryotes and is discussed first before the discussion of eukaryotes. Initiation takes place in a series of defined steps (Fig. 15-1). First, holoenzyme binds to the double-stranded promoter, forming what is called the closed complex. The specificity and strength of this interaction are dictated by sequence-specific contacts between the σ factor and the bases in the -10 and -35 elements of the promoter (Fig. 15-6). The second step in initiation is the formation of an open complex in which a 14-bp region around the transcription start site is unpaired producing a transcription bubble. This unpairing is accompanied by a conformational change in the polymerase that positions the single-strand DNA template in the active site and narrows the DNA-binding cleft, effectively closing the polymerase clamp. In the next step, the DNA template in the active site base-pairs with the first two ribonucleotides, and the first phosphodiester bond is catalyzed. This process is repeated until the nascent RNA reaches a length of eight to nine bases, at which point addition of bases to the growing RNA chain results in the unpairing of one base of the RNA-DNA hybrid, and the nascent RNA begins to exit through a channel on the surface of the polymerase. The resulting conformational change in polymerase leads to the release of σ factor and formation of a stable ternary (three-way) complex containing RNA polymerase, the DNA template, and the nascent RNA.
General Eukaryotic Transcription Factors
Purified eukaryotic RNA polymerase on its own cannot initiate transcription from promoters in vitro. Specific transcription can be obtained in vitro using extracts from nuclei, and fractionation of such extracts has led to the identification of additional factors necessary for specific transcription by purified RNA polymerase in vitro. Rather than a σ factor, eukaryotic RNA polymerases require multiple initiation factors. Most of these factors are unique to each RNA polymerase, and because they are required for transcription of most promoters (within each class), they are termed general transcription factors (GTFs). GTFs are remarkably conserved among different eukaryotes. Although most factors required for transcription by each class of polymerase are distinct, one of them, first identified as the TATA box–binding protein, participates in different protein complexes involved in each of the three polymerase systems. The next sections compare transcription by the three forms of eukaryotic RNA polymerase.
RNA Polymerase II Factors
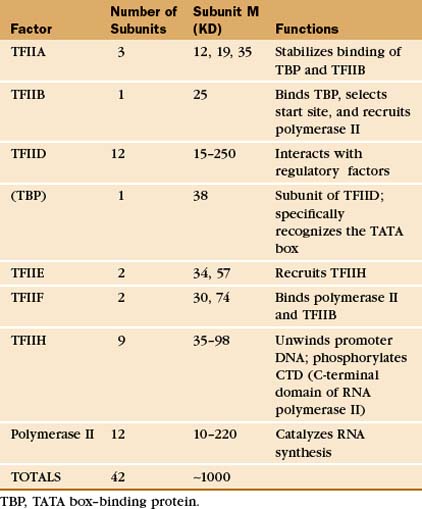
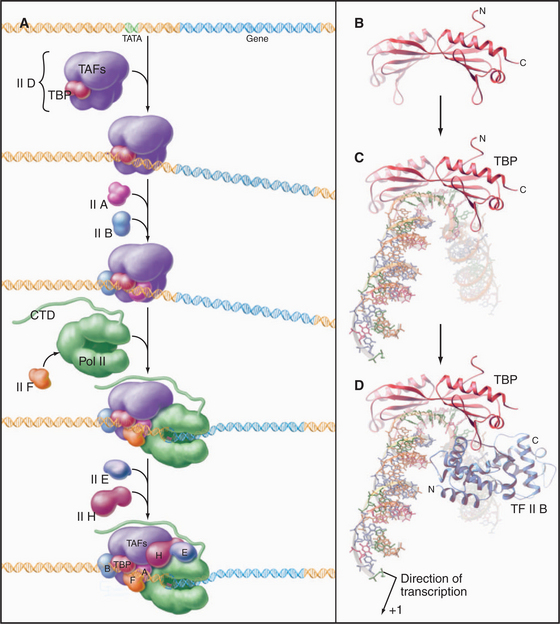
The RNA polymerase II GTFs comprise more than 20 polypeptides with an aggregate molecular weight of more than 106 D (Table 15-1). Before RNA polymerase II can initiate transcription in vitro, an ordered assembly of factors at the promoter must occur. Assembly of the RNA polymerase II preinitiation complex begins with the binding of TFIID, a large factor (˜700 kD) consisting of TATA box–binding protein (TBP) and a set of TBP-associated factors called TAFIIs (Fig. 15-7A). TBP alone is sufficient for basal transcription, while TAFs apparently serve as targets for further activation of transcription (see subsequent sections). TBP is the first polypeptide in the basal transcription machinery to recognize a specific DNA sequence during the initiation pro-cess. DNA binding is provided by a highly conserved C-terminal 180-amino-acid domain, which forms a saddle-shaped monomer with an axis of dyad symmetry (Fig. 15-7B). The underside of the TBP “saddle” binds to the minor groove of the TATA sequence, which is splayed open in the process. A pronounced DNA bend is produced at each end of the TATAAA element by the intercalation of phenylalanine side chains (Fig. 15-7C).
The TFIID-TATA box complex serves as a binding site for additional positive and negative regulators. TFIIA binding stabilizes the TBP-DNA interaction and prevents the binding of repressors that arrest further initiation complex formation.
The next step in assembly of the initiation complex is binding of TFIIB, which binds to one side of TBP and makes contacts with DNA upstream and downstream of the TATA box (Fig. 15-7D). Mutations in the yeast gene that encodes TFIIB show altered mRNA start-site selection, indicating that TFIIB establishes the spacing between the TATA box and the transcription start site. TFIIB interacts directly with TBP and RNA polymerase II and is thus essential for the next steps in initiation complex assembly.
RNA polymerase II enters into the preinitiation complex (see Fig. 15-7A) in association with TFIIF. This factor is related to bacterial σ factor and acts to stabilize the interaction of RNA polymerase II with TFIIB and TBP. In addition, TFIIF binds to free polymerase and prevents interactions with nonpromoter DNA sites.
TFIIH also contains a protein kinase that phosphorylates the CTD. This is Cdk-activating kinase, itself a Cdk-cyclin complex that phosphorylates and activates other cyclin-dependent kinases (see Fig. 40-14). In the initiation complex, phosphorylation of the CTD is thought to release it from interactions with GTFs and allow the transition to the transcription elongation phase. Other TFIIH subunits have been identified as components of the DNA repair machinery. Several genes encoding TFIIH subunits are mutated in the human DNA excision repair disease xeroderma pigmentosa, suggesting that TFIIH might serve to link transcription to DNA repair (see later section).
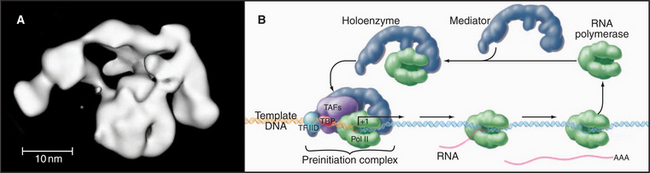
Mediator and the Holoenzyme
In vivo, many of the steps described previously involve the assembly of large macromolecular complexes containing RNA polymerase II, several of the GTFs, other factors that alter chromatin structure, and various additional transcription factors. One of these complexes, the mediator, contains over 20 polypeptides (many with unknown function) but lacks RNA polymerase II and the GTFs. Mediator reversibly interacts with RNA polymerase II and other factors to form a “holoenzyme,” which requires additional factors to be competent for initiation (Fig. 15-8). RNA polymerase II holoenzyme responds to transcription activators (de-scribed in a subsequent section) in vitro, suggesting that one role for the multitude of proteins in this complex is to offer multiple interaction sites for recruitment of holoenzyme to the promoter. Alternatively, a mediator lacking RNA polymerase II can be recruited to the promoter, where it subsequently attracts the polymerase. Thus, the mediator links DNA-bound activators to the basal transcription machinery. In this sense, the mediator acts as a coactivator. Other coactivators present in the holoenzyme act as chromatin remodeling factors (see subsequent section) that act to control access of the transcription machinery to the DNA template.