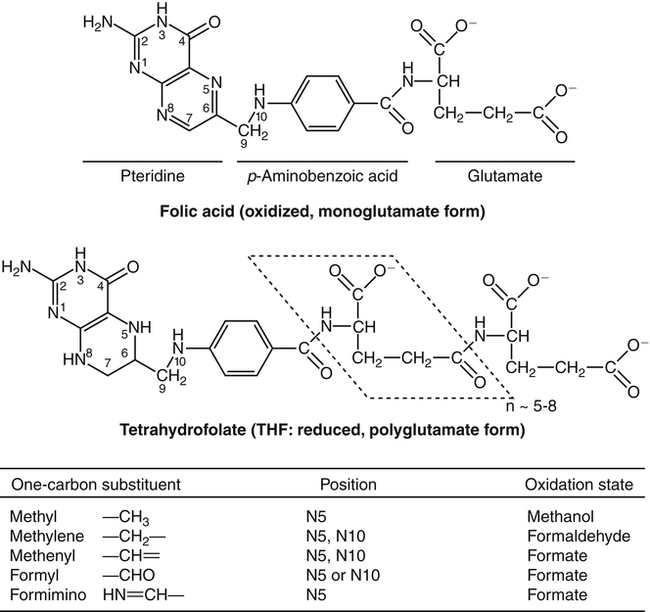
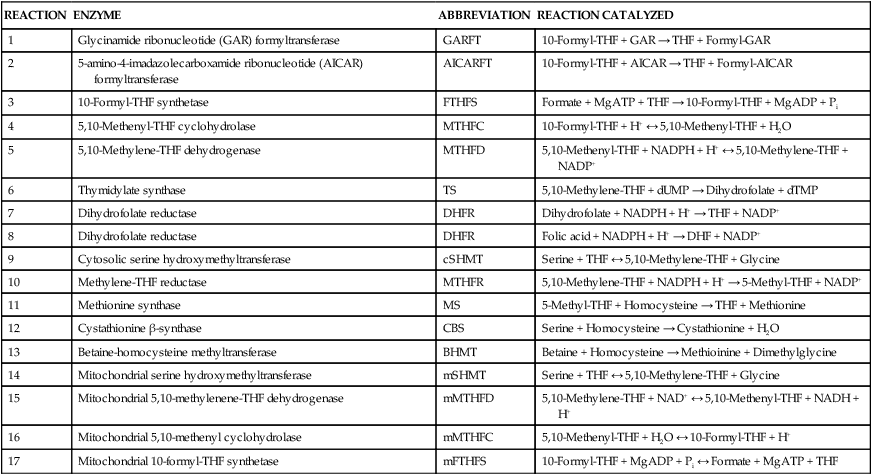
Marie A. Caudill, PhD, RD, Joshua W. Miller, PhD, Jesse F. Gregory, III, PhD; and Barry Shane, PhD The grouping of the nutrients folate, choline, vitamin B12, and vitamin B6 arises from their important roles as coenzymes, cosubstrates, or both in the one-carbon (1-C) metabolic network. Folate polyglutamates function in the transfer of 1-C units and are required for nucleotide biosynthesis and methionine reformation from homocysteine. Vitamin B6–dependent enzymes are intimately involved in the supply of 1-C units and in homocysteine catabolism. Vitamin B12 deficiency can cause a secondary deficiency of folate resulting in diminished nucleotide biosynthesis and cell division. Because choline serves as an alternative route to folate and vitamin B12–dependent homocysteine remethylation, there is an added layer of nutrient intermingling. Folate is the generic name for this water-soluble B vitamin. Common structural features of folate derivatives include (1) a pteridine bicyclic ring system, (2) p-aminobenzoic acid, and (3) one or more glutamic acid residues (Figure 25-1). Folic acid is the synthetic, monoglutamate, fully oxidized, and most stable form of the vitamin (see Figure 25-1). Folate metabolism involves the reduction of the pyrazine ring of the pterin moiety to the coenzymatically active tetrahydrofolate (THF) form, the elongation of the glutamate chain by the addition of L-glutamate residues in an unusual γ-peptide linkage, and the acquisition and oxidation or reduction of 1-C units at the N5 and/or N10 positions (see Figure 25-1). Most naturally occurring dietary folates are polyglutamate derivatives and must be hydrolyzed by a brush border membrane γ-glutamylhydrolase (glutamate carboxypeptidase II, GCPII) in the small intestine to monoglutamate forms before absorption across the intestinal mucosa. As folic acid is a monoglutamate, hydrolysis of glutamates by GCPII is not required. Absorption of folate monoglutamate is via a saturable carrier-mediated process, but a diffusion-like process also occurs at high folate concentrations. The main intestinal transporter, proton-coupled folate transporter (PCFT) encoded by the SLC46A1 gene, is a transmembrane protein that is highly expressed in enterocytes. PCFT belongs to the superfamily of solute carriers, functions optimally at low pH, and has a similar affinity for reduced folates and folic acid (Zhao et al., 2009a). Hereditary folate malabsorption is a consequence of loss-of-function mutations in the SLC46A1 gene. Bioavailability is a function of absorptive and metabolic processes that are influenced by many factors, including nutrient form, dietary intake, and individual genetic variation (McNulty and Pentieva, 2009; Caudill, 2010). In contrast to the high bioavailability of folic acid when given as a supplement or in fortified food, the bioavailability of naturally occurring food folate is variable and often incomplete. Several luminal factors can contribute to the lower bioavailability of food folate, including (1) entrapment of naturally occurring folates in the cellular structure or insoluble matrix of certain foods, (2) destruction of labile tetrahydrofolates during passage through the stomach, (3) inhibition of the intestinal deconjugation of polyglutamyl folates by food constituents, and (4) impairment of folate deconjugation by alteration of jejunal pH. Although pharmacological doses of folic acid are well absorbed, most of the vitamin is not retained in the body because tissues have a limited capacity to retain large amounts of folate. After folate absorption into the portal circulation (predominately 5-methyl-THF), much of this folate can be taken up by the liver via PCFT (Shane, 2009; Zhao et al., 2009a) (Figure 25-2). This folate–proton symporter is highly expressed on human liver basolateral membranes, which transport folates from the portal circulation. Liver and most, if not all, tissues also express the bidirectional reduced folate carrier (RFC), a product of the SLC19A1 gene (Zhao et al., 2009a). RFC is a transmembrane protein whose specificity for various folates differs among tissues and between the apical and basolateral membranes of cells. Affinities of RFC for reduced folates are in the low micromolar range, whereas affinities for folic acid are lower in the mid-micromolar range. Folate monoglutamates are exported from cells by RFC but also by select ATP-binding cassette (ABC) exporters such as the multidrug resistance (MDR)–associated proteins (Zhao et al., 2009a). 5-Methyl-THF is the predominate form of folate exported to plasma and is thus the circulatory form. Some folate is secreted in bile, but this can be reabsorbed in the intestine via an enterohepatic circulation. Folate receptors (FRα, FRβ, and FRγ), also referred to as folate-binding proteins, are a distinct class of folate transporters that are expressed by several peripheral tissues and display a high affinity for folic acid (in the nanomolar range). There are at least three distinct genes that code for the receptors, and the encoded protein is usually attached to the plasma membrane of cells via a glycosylphosphatidylinositol anchor (Zhao et al., 2009a). Internalization of folate occurs via a receptor-mediated endocytotic process. In the internalized endosome, folate is released from the receptor and is exported from the endosome into the cytosol by a mechanism that appears to be mediated by PCFT (Zhao et al., 2009b). Folate receptors are highly expressed in the choroid plexus, kidney proximal tubes, erythropoietic cells, and placenta, and in a number of human tumors. The presence of this high affinity transporter in the choroid plexus is thought to protect the brain from the effects of folate deficiency, as folate levels in the cerebrospinal fluid are considerably higher than in the peripheral circulation. A soluble form of folate-binding protein is also present at low levels in plasma and at high levels in human milk. More than 95% of tissue folates are polyglutamate species, primarily with chain lengths between five and eight glutamates. With most folate-dependent enzymes, the polyglutamates usually exhibit greatly increased affinities for these enzymes and are more effective than monoglutamates as substrates. The polyanionic nature of the polyglutamate chain, coupled with binding of folate polyglutamates by intracellular proteins, allows tissues to retain and concentrate polyglutamate forms of the vitamin. Folate coenzymes are found primarily in the mitochondria and cytosol of the cell, and accumulation of folate in these compartments requires the conversion of folates to polyglutamates, which is catalyzed by the enzyme folylpolyglutamate synthetase (Shane, 2009): Tissues contain a soluble lysosomal γ-glutamylhydrolase activity, sometimes called folate conjugase, which is involved in the hydrolysis of polyglutamates with their subsequent release from the tissue in the monoglutamate form. However, the major route of folate turnover and catabolism appears to involve the degradation of folate coenzymes to pterin derivatives and aminobenzoylpolyglutamates via oxidative cleavage at the C9, N10 bond (see Figure 25-1). Methenyl-THF synthetase, the normal enzymatic function of which is the conversion of 5-formyl-THF to 5,10-methenyl-THF, is responsible for some of this cleavage. The aminobenzoylpolyglutamates generated are hydrolyzed to aminobenzoylglutamate by lysosomal γ-glutamylhydrolase and are partly N-acetylated, at least in liver, and the N-acetyl-aminobenzoylglutamate is excreted in the urine. In human populations, only small amounts of intact folate are usually found in urine (except in populations exposed to folic acid fortification and/or consuming supplemental folate), and cleavage products represent the bulk of the excretion. Under normal conditions of dietary intake and status, whole-body folate stores turn over slowly with a half-life in excess of 100 days. Folate coenzymes act as acceptors or donors of 1-C units in a variety of reactions involved in nucleotide biosynthesis and methyl metabolism in mammalian tissues. Methionine, thymidylate, and purine synthesis are the major pathways of 1-C metabolism and occur in the cytosol and nucleus (see Figure 25-2). These biosynthetic cycles are interconnected, and interconversion of the folate coenzymes is mediated by the trifunctional enzyme, C1-THF synthase. Consequently, folate metabolism and its regulation are interwoven, and factors that regulate any one cycle of 1-C metabolism will influence folate availability for the other cycles. Extensive folate metabolism also occurs in the mitochondria, where mitochondrial folate metabolism plays an important role in the provision of 1-C units for cytosolic 1-C metabolism. The C-8 and C-2 positions of the purine ring are derived from 10-formyl-THF in reactions catalyzed by glycinamide ribonucleotide formyltransferase (GARFT) (see reaction 1 in Figure 25-2 and Table 25-1) and 5-amino-4-imidazolecarboxamide ribonucleotide formyltransferase (AICARFT) (see reaction 2 in Figure 25-2 and Table 25-1). 10-Formyl-THF is formed by the formylation of THF in a reaction catalyzed by 10-formyl-THF synthetase (see reaction 3 in Figure 25-2 and Table 25-1), a step that represents the primary entry point of 1-C units for cytosolic folate-dependent biosynthetic reactions (Stover, 2009). TABLE 25-1 Major Metabolic Reactions for Folate* *As shown in Figure 25-2. Reactions 3, 4, and 5 represent activities of the cytosolic trifunctional enzyme, C1-THF synthase. 10-Formyl-THF synthetase activity resides on the C-terminal domain of the trifunctional enzyme, C1-THF synthase, which is encoded by the MTHFD1 gene. The N-terminal domain of C1-THF synthase contains cyclohydrolase and dehydrogenase activities, which enable the reversible interconversions of 10-formyl-THF to 5,10-methenyl-THF (see reaction 4 in Figure 25-2 and Table 25-1) and 5,10-methenyl-THF to 5,10-methylene-THF (see reaction 5 in Figure 25-2 and Table 25-1). Because of the reversibility of these reactions, 10-formyl-THF can also be obtained by the oxidation of 5,10-methylene-THF. In vivo, however, this pathway is driven in the reductive direction (i.e., toward 5,10-methylene-THF) by the higher ratio of NADPH to NADP+ in the cytosol of cells. Folate, as 5,10-methylene-THF, is required for the synthesis of the pyrimidine, thymidylate. Thymidylate synthase catalyzes the transfer of formaldehyde from folate to the 5-position of deoxyuridine monophosphate (dUMP) for the formation of deoxythymidine monophosphate (dTMP) (see reaction 6 in Figure 25-2 and Table 25-1). The THF of 5,10-methylene-THF provides the reducing component for reduction of the transferred methylene moiety to a methyl group of dTMP and is oxidized to DHF in the process. DHF is inactive as a coenzyme and has to be reduced back to THF, in a reaction catalyzed by DHF reductase, before it can play a further role in 1-C metabolism (see reaction 7 in Figure 25-2 and Table 25-1). The major role of DHF reductase is the reduction of DHF formed during thymidylate synthesis to THF. Dihydrofolate reductase also catalyzes the reduction of synthetic folic acid to DHF (see reaction 8 in Figure 25-2 and Table 25-1) and then THF; however, folic acid is a poorer substrate than DHF (Bailey and Ayling, 2009). 5,10-Methylene-THF is generated from 10-formyl-THF through the cyclohydrolase and dehydrogenase activities of the trifunctional protein, C1-THF synthase, as previously described. Alternatively, 5,10-methylene-THF is obtained by cytosolic serine hydroxymethyltransferase (cSHMT), a pyridoxal 5′-phosphate (PLP)-dependent enzyme that catalyzes the reversible transfer of formaldehyde from serine to THF to generate 5,10-methylene-THF and glycine (see reaction 9 in Figure 25-2 and Table 25-1). Mammalian cells have two genes that encode SHMT isozymes. These isozymes share 63% amino acid sequence identity and are encoded by SHMT1 and SHMT2 (Stover, 2009). SHMT1 encodes the cytosolic isozyme, and SHMT2 encodes the mitochondrial isozyme found in mature cells. However, SHMT2 has recently been shown to encode two transcripts, one for SHMT2 (mitochondrial) and one for SHMT2α. The transcript for SHMT2α lacks exon 1 and encodes a protein that is found in the cytosol and nucleus during the S (DNA synthesis) phase of the cell cycle (see next paragraph) (Anderson and Stover, 2009). The C3 of serine, a nonessential amino acid derived from glucose or the diet, is a major source of 1-C units for folate metabolism that mainly enter cytosolic folate-mediated 1-C metabolism as formate (Davis et al., 2004). The three enzymes that constitute the de novo thymidylate synthesis pathway in mammals—cytosolic serine hydroxymethyltransferase (SHMT1 and SHMT2α), thymidylate synthase, and DHF reductase—undergo sumoylation and nuclear import during the S phase of the cell cycle (Anderson et al., 2007; Woeller et al., 2007). Thymidylate synthase activity is expressed only in replicating tissues, and expression of thymidylate synthase and DHF reductase mRNA is highest during the S phase of the cell cycle. Compartmentation of the thymidylate biosynthetic enzymes in the nucleus may allow for folate-dependent deoxythymidine triphosphate (dTTP) biosynthesis directly at the replication fork (Anderson and Stover, 2009). A major cytosolic cycle of 1-C incorporation involves the reduction of 5,10-methylene-THF to 5-methyl-THF, followed by the transfer of the methyl group to homocysteine to form methionine and regenerate THF. 5,10-Methylene-THF reduction is catalyzed by the flavoprotein methylenetetrahydrofolate reductase (MTHFR) (see reaction 10 in Figure 25-2 and Table 25-1). The reaction is irreversible under in vivo conditions and is the committed step in the flux of 1-C units to the methionine resynthesis cycle. The next enzyme in this cycle, methionine synthase, is one of only two vitamin B12–dependent mammalian enzymes, and it catalyzes the transfer of the methyl group from 5-methyl-THF to homocysteine (see reaction 11 in Figure 25-2 and Table 25-1). The methionine synthase reaction is the only reaction in which the methyl group of 5-methyl-THF can be metabolized in mammalian tissues. Although methionine is an essential amino acid, the methionine synthase reaction plays a major role in methyl group metabolism because it allows the reuse of the homocysteine backbone as a carrier of methyl groups derived primarily from the C3 of serine. The enzyme contains tightly bound cob(I)alamin, and the reaction proceeds via a methylcob(III)alamin intermediate as described in the vitamin B12 section of this chapter. Homocysteine is not found in the diet but arises from hydrolysis of S-adenosylhomocysteine (AdoHcy), the product of S-adenosylmethionine (AdoMet)–dependent methylation reactions. Homocysteine can be metabolized to cysteine in reactions catalyzed by two PLP-dependent enzymes, cystathionine β-synthase (see reaction 12 in Figure 25-2 and Table 25-1) and cystathionine γ-lyase. Alternatively, homocysteine can be converted back to methionine via the folate-dependent methionine synthase reaction (see reaction 11 in Figure 25-2) or by betaine–homocysteine methyltransferase, which catalyzes the transfer of one of the methyl groups of betaine to homocysteine to generate methionine and dimethylglycine (see reaction 13 in Figure 25-2 and Table 25-1). The extent of homocysteine remethylation or transsulfuration is tissue-dependent, and many tissues export homocysteine and cystathionine into the circulation. Tissue levels of homocysteine are normally low; elevated homocysteine levels increase AdoHcy levels, an inhibitor of many methylation reactions. Kidney and liver are thought to be important organs for homocysteine remethylation and for transsulfuration. Remethylation is also dependent on the methyl group status of the tissue. The major regulator of the folate-dependent methionine cycle is AdoMet, which is a potent allosteric inhibitor of MTHFR. Liver contains a high Km AdoMet synthetase, and hepatic levels of AdoMet reflect methionine status. High levels of AdoMet inhibit MTHFR, reducing synthesis of 5-methyl-THF and hence remethylation of homocysteine. At the same time, the high levels of AdoMet activate cystathionine β-synthase, stimulating transsulfuration of homocysteine to cysteine. Conversely, when AdoMet is low, remethylation of homocysteine is favored and transsulfuration is inhibited. MTHFR activity is also modified by the well-described genetic variant, 677C→T within this gene, which reduces enzyme activity and modifies disease risk (Christensen and Rozen, 2009). Mitochondrial folate-mediated 1-C metabolism generates 1-C units in the form of formate through the catabolism of serine, glycine, and choline. Redox and equilibrium conditions support the flow of 1-C units through the mitochondria in the oxidative direction of 5,10-methylene-THF to 10-formyl-THF to formate (Tibbetts and Appling, 2010); however, many questions remain regarding the control of 1-C flux through the mitochondria, which may be tissue and species specific. Reduced monoglutamate coenzyme forms, likely the monoglutamate of THF or 5-formyl-THF, are transported across the mitochondrial inner membrane by the mitochondrial folate transporter (MFT) (Tibbetts and Appling, 2010; Titus and Moran, 2000) and then undergo polyglutamation. Entry of serine, glycine, and other 1-C donors into the mitochondria occurs via carrier-mediated processes, although the proteins involved in these transport processes are not well defined. Mitochondrial PLP-dependent serine hydroxymethyltransferase (mSHMT) catalyzes the reversible transfer of serine C3 to THF to form glycine and 5,10-methylene-THF (see reaction 14 in Figure 25-2 and Table 25-1). 5,10-Methylene-THF can also be produced by the transfer of 1-C units to THF from dimethylglycine, sarcosine, and glycine. The dehydrogenase and cyclohydrolase activities of the mitochondrial bifunctional C1-THF synthase catalyze the reversible conversions of 5,10-methylene-THF to 5,10-methenyl-THF, and 5,10-methenyl-THF to 10-formyl-THF (see reactions 15 and 16, respectively, in Figure 25-2 and Table 25-1). The bifunctional C1-THF synthase encoded by the MTHFD2 gene is expressed in mammalian embryos (Christensen and Mackenzie, 2008), whereas the enzyme encoded by the putative MTHFD2L gene is expressed in mammalian adults (Tibbetts and Appling, 2010). It is hypothesized that MTHFD2L expression replaces MTHFD2 as development progresses, thereby maintaining mitochondrial dehydrogenase/cyclohydrolase activities throughout development and adulthood (Tibbetts and Appling, 2010). A separate gene, MTHFD1L, encodes a monofunctional 10-formyl-THF synthetase for mitochondrial formate production and THF regeneration (see reaction 17 in Figure 25-2 and Table 25-1). Efflux of formate into the cytosol enables its use in cytosolic folate mediated 1-C metabolism. The 1-C units can also be disposed of by conversion to CO2 in a reaction catalyzed by 10-formyl-THF dehydrogenase: TABLE 25-2 Major Metabolic Reactions for Choline* Alternatively, the 1-C can be used for mitochondrial protein synthesis whereby the initiator methionyl-tRNA (Met-tRNAfMet) is formylated to formylmethionyl-tRNA (fMet-tRNAfMet) (Li et al., 2000) in a reaction catalyzed by methionyl-tRNA formyltransferase: In addition to 1-Cs derived from mitochondrial and cytosolic serine catabolism, 1-C units are also derived from histidine and purine degradation (Stover, 2009). 1-C units derived from histidine and purines enter cytosolic folate-mediated metabolism as 5,10-methenyl-THF, which exists in equilibrium with 10-formyl-THF. The C2 of the imidazole ring of histidine provides 1-C units at the oxidation level of formate. Cytosolic formiminoglutamate formiminotransferase catalyzes the transfer of a formimino group from formiminoglutamate, an intermediate in the histidine catabolism pathway, to THF:
Folate, Choline, Vitamin B12, and Vitamin B6
Chemistry of Folate
Folate Absorption
Folate Bioavailability
Transport of Folate
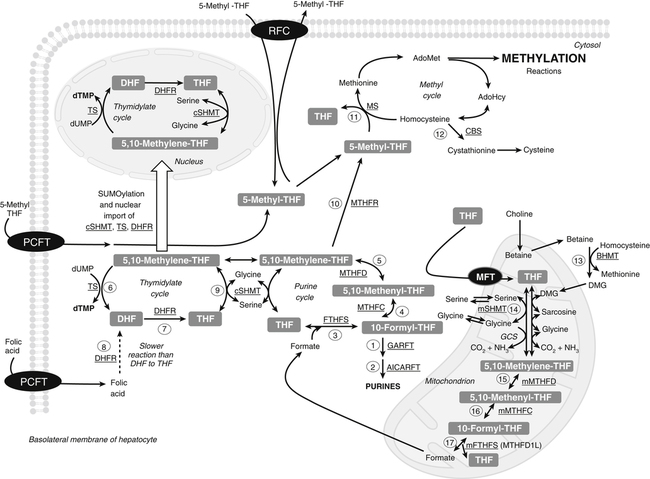
Tissue Accumulation of Folate
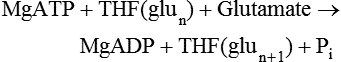
Folate Turnover
Metabolic Functions of Folate
Nucleotide Synthesis
Purine Cycle
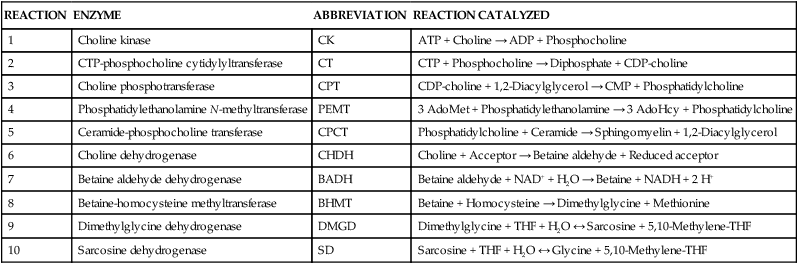
REACTION
ENZYME
ABBREVIATION
REACTION CATALYZED
1
Glycinamide ribonucleotide (GAR) formyltransferase
GARFT
10-Formyl-THF + GAR → THF + Formyl-GAR
2
5-amino-4-imadazolecarboxamide ribonucleotide (AICAR) formyltransferase
AICARFT
10-Formyl-THF + AICAR → THF + Formyl-AICAR
3
10-Formyl-THF synthetase
FTHFS
Formate + MgATP + THF → 10-Formyl-THF + MgADP + Pi
4
5,10-Methenyl-THF cyclohydrolase
MTHFC
10-Formyl-THF + H+ ↔ 5,10-Methenyl-THF + H2O
5
5,10-Methylene-THF dehydrogenase
MTHFD
5,10-Methenyl-THF + NADPH + H+ ↔ 5,10-Methylene-THF + NADP+
6
Thymidylate synthase
TS
5,10-Methylene-THF + dUMP → Dihydrofolate + dTMP
7
Dihydrofolate reductase
DHFR
Dihydrofolate + NADPH + H+ → THF + NADP+
8
Dihydrofolate reductase
DHFR
Folic acid + NADPH + H+ → DHF + NADP+
9
Cytosolic serine hydroxymethyltransferase
cSHMT
Serine + THF ↔ 5,10-Methylene-THF + Glycine
10
Methylene-THF reductase
MTHFR
5,10-Methylene-THF + NADPH + H+ → 5-Methyl-THF + NADP+
11
Methionine synthase
MS
5-Methyl-THF + Homocysteine → THF + Methionine
12
Cystathionine β-synthase
CBS
Serine + Homocysteine → Cystathionine + H2O
13
Betaine-homocysteine methyltransferase
BHMT
Betaine + Homocysteine → Methioinine + Dimethylglycine
14
Mitochondrial serine hydroxymethyltransferase
mSHMT
Serine + THF ↔ 5,10-Methylene-THF + Glycine
15
Mitochondrial 5,10-methylenene-THF dehydrogenase
mMTHFD
5,10-Methylene-THF + NAD+ ↔ 5,10-Methenyl-THF + NADH + H+
16
Mitochondrial 5,10-methenyl cyclohydrolase
mMTHFC
5,10-Methenyl-THF + H2O ↔ 10-Formyl-THF + H+
17
Mitochondrial 10-formyl-THF synthetase
mFTHFS
10-Formyl-THF + MgADP + Pi ↔ Formate + MgATP + THF
Thymidylate Cycle
Methyl Metabolism
Methionine Cycle
Sources of 1-C Units for Biosynthetic Reactions
Mitochondrial Generation of 1-C Units
REACTION
ENZYME
ABBREVIATION
REACTION CATALYZED
1
Choline kinase
CK
ATP + Choline → ADP + Phosphocholine
2
CTP-phosphocholine cytidylyltransferase
CT
CTP + Phosphocholine → Diphosphate + CDP-choline
3
Choline phosphotransferase
CPT
CDP-choline + 1,2-Diacylglycerol → CMP + Phosphatidylcholine
4
Phosphatidylethanolamine N-methyltransferase
PEMT
3 AdoMet + Phosphatidylethanolamine → 3 AdoHcy + Phosphatidylcholine
5
Ceramide-phosphocholine transferase
CPCT
Phosphatidylcholine + Ceramide → Sphingomyelin + 1,2-Diacylglycerol
6
Choline dehydrogenase
CHDH
Choline + Acceptor → Betaine aldehyde + Reduced acceptor
7
Betaine aldehyde dehydrogenase
BADH
Betaine aldehyde + NAD+ + H2O → Betaine + NADH + 2 H+
8
Betaine-homocysteine methyltransferase
BHMT
Betaine + Homocysteine → Dimethylglycine + Methionine
9
Dimethylglycine dehydrogenase
DMGD
Dimethylglycine + THF + H2O ↔ Sarcosine + 5,10-Methylene-THF
10
Sarcosine dehydrogenase
SD
Sarcosine + THF + H2O ↔ Glycine + 5,10-Methylene-THF
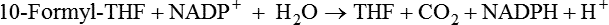

Cytosolic Generation of 1-C Units
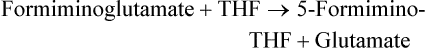
![]()
Stay updated, free articles. Join our Telegram channel

Full access? Get Clinical Tree


Folate, Choline, Vitamin B12 , and Vitamin B6