Foamy Viruses
Axel Rethwilm
Dirk Lindemann
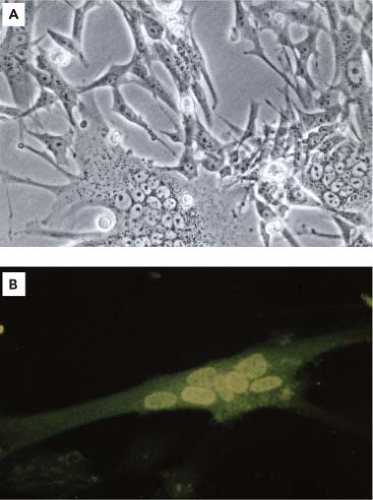
The name foamy virus (FV) was coined in the 1950s to acknowledge the spontaneous emergence of the typical foamy cytopathic effect (CPE) produced in response to FV infection (Fig. 52.1A). This is characterized by multinucleated syncytia and vacuolation in primary monkey kidney cultures leading to a “foamy” appearance.47,154,186,206 Subsequently, it was found that CPE development was attributed to the fact that the monkeys were latently infected with a transmittable agent. Following the discovery of reverse transcriptase, the transmitted agent was shown to be a retrovirus.172 The first 30 years of FV research dealt mainly with the identification of infected monkeys, prior to sacrifice, as sources of primary cell cultures. Molecular cloning of the first FV—at that time believed to be a human isolate—permitted functional studies on the replication of FVs.192 FV research gained momentum following the discovery that these viruses replicate differently from all other retroviruses.133,264 This culminated in the finding that the FV infectious genome appears to be DNA rather than RNA.160,203,269 In brief, the FV replication strategy combines those of retroviruses with some characteristics of hepadnaviruses, such as hepatitis B virus (HBV), with other properties that are unique to FVs.120,186,190 In virtually any aspect that has been examined, FVs replicate differently from all other retroviruses. This has led to the definition of two retroviral subfamilies, the Orthoretrovirinae, which encompass all retrovirus genera except FVs, and the Spumaretrovirinae, which constitute only the spumaviruses or FVs.134 This chapter will summarize what is known about the biology of these viruses; for reviews on particular aspects, such as FV vectors, the reader is referred to more specialized reviews.132,188,246,249 We shall focus on FVs of primates and mention the nonprimate viruses only when they become relevant.
Foamy Virus Isolation and Diagnosis of Infection
FV isolation is relatively easy on primary fibroblasts from throat swabs, although any other tissue as source of virus can be used.94,154,166,186 Virus isolation from peripheral blood lymphocytes is greatly enhanced by the addition of anti–γ-interferon (γ-IFN) antibodies.56 A virus isolate displaying the typical giant cells’ CPE (Fig. 52.1A) can be confirmed by the detection of reverse transcriptase activity, by immunofluorescence assay (IFA) demonstrating a predominantly nuclear antigen (Fig. 52.1B), or by nucleic acid detection methods.94,154,166,186,222
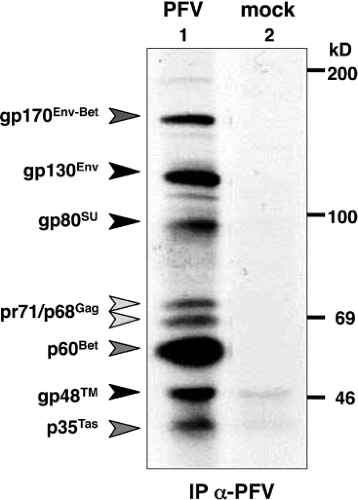
The diagnosis of an FV infection can be made by demonstrating antibodies against the main structural proteins (typically the Gag doublet, see later discussion) in serologic assays,71 such as immunoblots or radioimmunoprecipitation assays (Fig. 52.2). The choice of the right antigen in these assays is important, because the reaction is, at least in part, virus type specific.96,102,112 To verify an FV infection, the serologic analysis should be combined with nucleic acid detection methods (typically polymerase chain reaction [PCR]). For this, the amplification of a conserved (∼420 bp) fragment from the integrase (IN) domain of the pol gene has proven to be extremely useful.220
Natural History and Trans-Species Transmissions
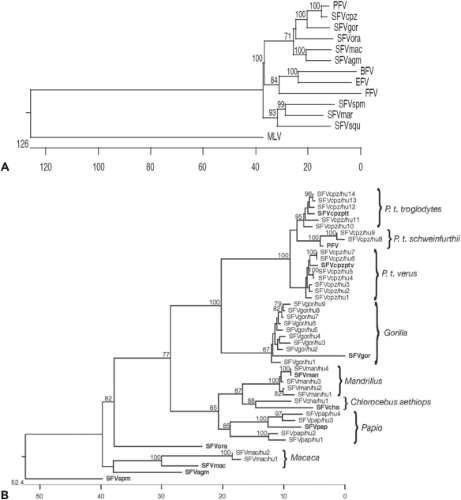
Various vertebrate species are naturally infected with FVs. Table 52.1 and Figure 52.3A give an overview of some isolates.
In addition to FVs of Great Apes,94,95,154,186 Old and New World simians,94,154,186 and prosimians,99,154,186 FV infections appear to occur worldwide in bovines and other Artiodactyla,5,59,148,149a,154,186 equines, 119,154,186,242 and felidae.154,186,196 Whether sea mammals are natural hosts has not been intensively investigated.110,154,186 Probably all monkey species harbor an FV.94,100,135,154,186 Prevalence in the natural host in the wild may be as high as 100% and is usually more than 30%.94,135,154,186 It is therefore not unlikely that FVs are, in terms of prevalence, the most successful of all retroviruses. Although host restriction factors show some species restriction (see later discussion), FVs have been reported to cross the species barrier between monkeys and apes in captivity or in the wild.94,124,135 In their hosts, FVs cause lifelong persistent infections of a benign nature, often in the presence of neutralizing antibodies.94,154,186 Laboratory animals, such as mice and rabbits, have also been infected in the absence of overt disease.26,93,94,208,214
In addition to FVs of Great Apes,94,95,154,186 Old and New World simians,94,154,186 and prosimians,99,154,186 FV infections appear to occur worldwide in bovines and other Artiodactyla,5,59,148,149a,154,186 equines, 119,154,186,242 and felidae.154,186,196 Whether sea mammals are natural hosts has not been intensively investigated.110,154,186 Probably all monkey species harbor an FV.94,100,135,154,186 Prevalence in the natural host in the wild may be as high as 100% and is usually more than 30%.94,135,154,186 It is therefore not unlikely that FVs are, in terms of prevalence, the most successful of all retroviruses. Although host restriction factors show some species restriction (see later discussion), FVs have been reported to cross the species barrier between monkeys and apes in captivity or in the wild.94,124,135 In their hosts, FVs cause lifelong persistent infections of a benign nature, often in the presence of neutralizing antibodies.94,154,186 Laboratory animals, such as mice and rabbits, have also been infected in the absence of overt disease.26,93,94,208,214
Humans are not a natural host of FVs.186 Indeed, the best-studied FV was once believed to be of human origin.1,143 It has now been designated as the prototype foamy virus (PFV).187 Initial reports on naturally occurring human infections144,147 were not confirmed by the large-scale screening of more than 5,000 samples using sophisticated methods such as a combination of antibody detection and PCR.2,222 Furthermore, postulated associations of FVs with human disease states, mainly autoimmune disorders and neurologic diseases of unknown origin could not be validated.40,86,201,223 Because of the close nucleotide sequence homology to FVs from the Pan troglodytes schweinfurthii chimpanzee sub-species,88,135 the single human isolate from a Kenyan patient is now believed to have resulted from a trans-species transmission of a virus from these chimpanzees that were once more prevalent in East Africa.51 Primate FVs are currently not circulating in the human population. However, humans are susceptible to zoonotic transmissions of nonhuman primate (NHP) FVs.218,232 Altogether, around 100 human infections with NHP FVs have been confirmed worldwide. In a survey, the Centers for Disease Control and Prevention identified approximately 2% seropositives and virus DNA positives among several hundred samples from occupationally to NHP-exposed persons and could isolate virus in several instances.22,87 Some of these
infections date back decades and were the result of severe monkey bites.22,87 FV infection has also been identified in African bushmeat hunters.30 Other persons at risk are those living in close proximity to quasi-wild NHPs (e.g., at Asian temple sites) or individuals possessing an NHP pet.101 A very special case may be human recipients of NHP xenotransplants.4 Human infections are lifelong; however, none induced any disease and remained unrecognized prior to the investigation. The viral load in buffy-coat cells of infected humans revealed a very low number of FV DNA copies.30 Moreover, in contrast to lentiviruses, in vivo adaptation to what can be called a human FV has not occurred. Even after decades of infection, FVs in humans have remained relatively unchanged at the nucleotide level, and the transmitting donor species can be readily identified22,87 (i.e., a baboon FV will remain a baboon FV even after decades in a human host) (Fig. 52.3B). Although the sample size is relatively small, there is no indication of human-to-human transmission even between close contacts.22,64a Because FV can be transmitted by transfusion,25,111 infected persons have been advised not to donate blood to avoid new human retrovirus infections.85
infections date back decades and were the result of severe monkey bites.22,87 FV infection has also been identified in African bushmeat hunters.30 Other persons at risk are those living in close proximity to quasi-wild NHPs (e.g., at Asian temple sites) or individuals possessing an NHP pet.101 A very special case may be human recipients of NHP xenotransplants.4 Human infections are lifelong; however, none induced any disease and remained unrecognized prior to the investigation. The viral load in buffy-coat cells of infected humans revealed a very low number of FV DNA copies.30 Moreover, in contrast to lentiviruses, in vivo adaptation to what can be called a human FV has not occurred. Even after decades of infection, FVs in humans have remained relatively unchanged at the nucleotide level, and the transmitting donor species can be readily identified22,87 (i.e., a baboon FV will remain a baboon FV even after decades in a human host) (Fig. 52.3B). Although the sample size is relatively small, there is no indication of human-to-human transmission even between close contacts.22,64a Because FV can be transmitted by transfusion,25,111 infected persons have been advised not to donate blood to avoid new human retrovirus infections.85
Table 52.1 Examples of Foamy Virus Isolates from Different Species of Primate and Nonprimate Origins | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
For reasons that are not fully understood, men appear to be a “dead-end” host for primate FVs. With respect to human infections by non-NHP FVs, it is worth mentioning that the antibody screening of more than 200 veterinarians at risk of acquiring a feline foamy virus (FFV) infection did not reveal a single positive case.28 Based on this result, it appears unlikely that bovine or equine foamy virus (BFV and EFV, respectively) can infect humans. However, this has not been thoroughly investigated.
Evolution of Foamy Viruses
Exogenous FVs are very ancient retroviruses. Exogenous FVs are very ancient retroviruses. There exist three examples of endogenous FVs, which suggest that exogenous FVs were around before that times: 1) Katzourakis et al.108 reported the detection of endogenous FVs (SloEFV) in the South American sloths genome (Choloepus hoffmanni), 2) Han and Worobey71a described endogenous FVs (PSFVaye) in the Madagascan aye-aye, a primitive lemur species (Daubentonia madagascarensis), and more surprinsingly, 3) in the latimera (Coelacanth) genome71b (CoeEFV), suggesting that exogenous FVs existed more than 400 million years ago in species outside the mammals. Moreover, once an exogenous FV has adapted to its host, it mutates only slightly faster than the host mitochondrial DNA.234 The substitution rate of simian foamy viruses (SFVs) has been estimated to be around 1.7 × 10−8 per site and year.234 This makes FVs the most genetically stable of viruses, with an RNA phase in replication. For instance, the FV mutation rate is approximately ten times lower than primate T-cell lymphotropic virus type 1 (PTLV-1), a virus that replicates primarily by a proviral expansion mechanism (i.e., through DNA) (see Chapter 48).
FVs combine two favorable features: They are of extraordinary genetic stability and, if not acquired by trans-species transmissions, always point to the host species from which they were derived, and they are shed in feces. Owing to these features, NHP phylogeographic and conservational issues can be addressed easily without animal disturbance.135,220
Curiously, the fidelity of the reverse transcriptase enzyme (RT) does not reflect this enormous genetic stability. If analyzed in vitro, the PFV RT was found to be of exceptional high processivity but of low fidelity.23,24,197 The mutation rate of PFV RT (approximately 1.7 × 10−4 per site and replication round) is similar to that of human immunodeficiency virus (HIV).24 Most mutations found were small deletions and insertions.24 If analyzed in cell culture, such deletions and insertions were not found. However, a point mutation error rate of 1.1 × 10−5 per site and round of replication remained.62
Thus, the genetic stability of FVs at the molecular level is not currently understood, and the involvement of as yet unidentified specific cellular factor(s) may play a role in this process.
Replication In Vitro
The host cell range for FVs is quite broad and includes species-independent primary cells or cell lines of fibroblastoid, epithelial, and lymphoblastoid origin, such as various B and T lymphocytes, and cells of erythroid and of myeloid lineages.94,154,159,186,268 Upon replication in adherent cell cultures, FVs induce massive multinucleated giant cell CPE (Fig. 52.1A), and apoptosis is thought to be the ultimate cause of cell death.158 Vacuolization of cells is often only observed using primary isolates.
The paucity of cell lines resistant to FV infections has hindered the identification of the cellular receptor(s) required for entry by classical approaches. It is now appreciated that all FVs use the same cellular receptor, including those present on bird, reptile, and fish cells.12,89,230 Only two cell lines—Pac-2 zebrafish embryonic fibroblasts and the G1E-ER4 human erythroid precursors—have been reported to be refractory to infection.230 Thus, the means to screen complementary DNA (cDNA) libraries for FV receptor–related genes has now been established.
Whereas the characteristic CPE develops in adherent cells, this hallmark of FVs is often absent in cells of lymphoblastoid origin, in which FVs appear to become latent and intermittently reactivatable. Latently infected cells do not undergo syncytium formation and death but proliferate with normal kinetics and produce low amounts of virus.268 As judged from Southern blots, the viral DNA copy number oscillated in infected lymphocytes (unpublished observation). Interestingly, chemical treatment of lymphocytes (e.g., with phorbol esters) may induce the latent virus and cause cell death owing to activated viral replication.157,268 Although this is reminiscent of the lymphotropic herpesviruses, in FVs the molecular basis for virus reactivation has not been investigated nor have sites within the cellular or FV DNA genome responsive to the drug-mediated reactivation been mapped. Whether the methylation of FV DNA that has been observed in a cell culture model219 contributes to in vivo latency remains unresolved, because there is no evidence of transcriptional down-regulation of FV vectors by methylation following their introduction in vivo.90,169,225
Replication in the Natural Host In Vivo
Recently, Liu et al135 used methods similar to those employed to demonstrate that human immunodeficiency virus type 1 (HIV-1) was derived from simian immunodeficiency virus from chimpanzees (SIVcpz; i.e., they collected and analyzed fecal samples from wild chimpanzees; see Chapters 49 and 50). They found that simian foamy virus from chimpanzees (SFVcpz) is widely distributed among wild chimpanzees with a
phylogeographic distribution and is transmitted horizontally, because babies younger than 2 years were free from FVs and infection rates increase with age. Moreover, they determined that superinfection by SFVs from lower primates and frequent recombination events occur. The most interesting finding by Liu et al has been the detection of viral RNA but not viral DNA in the fecal samples.135 This finding directly relates to the FV replication pathway (see later discussion). However, Liu et al have not investigated whether the RNA-containing virus transmits the infection, and it is not known in which cell type these viruses were produced. It is possible that the DNA content in the fecal samples may have been too low to be detected even with very sensitive methods.
phylogeographic distribution and is transmitted horizontally, because babies younger than 2 years were free from FVs and infection rates increase with age. Moreover, they determined that superinfection by SFVs from lower primates and frequent recombination events occur. The most interesting finding by Liu et al has been the detection of viral RNA but not viral DNA in the fecal samples.135 This finding directly relates to the FV replication pathway (see later discussion). However, Liu et al have not investigated whether the RNA-containing virus transmits the infection, and it is not known in which cell type these viruses were produced. It is possible that the DNA content in the fecal samples may have been too low to be detected even with very sensitive methods.
The tissue distribution of SFV and sites of in vivo replication have been investigated using another approach. As expected from the broad host cell range of FVs seen in vitro, viral DNA was detected at a frequency of one genome copy per 102 to 103 cells in every organ examined.54 Because animals were perfused prior to the analysis of nucleic acids, infected lymphocytes were not detected, although these cells were the probable vehicles of virus dissemination in vivo.54,55 As judged from in situ hybridization experiments, viral RNA, indicative of active virus replication, was confined to superficial cells of the oral mucosa.54,165 Thus, it appears that only cells, which are destined to be shed, are productively infected and undergo lytic replication in vivo. Differential expression of yet undisclosed host factors restricting viral replication in other tissues is a likely explanation for this observation.
It is generally assumed that FVs are transmitted among NHPs through saliva via social contacts, including aggressive activities such as biting among young animals.31,135 These contacts and lactation are also suspected to be the main transmission route of the FFV,259 whereas BFV probably is mainly transmitted via milk from infected cows to offspring.198
Early studies suggested that drug-induced immunosuppression of infected African green monkeys did not result in SFV-related symptoms yet enhanced the frequency of virus isolation (D. Neumann-Haefelin, Freiburg, Germany, personal communication). It was subsequently found that in dually SFV- and simian immunodeficiency virus (SIV)-infected and severely immunosuppressed macaques, the predominant site of FV replication changed from the oral mucosa to the small intestine.164 However, SFV-related diseases did not occur. This was also observed in cats dually infected with feline immunodeficiency virus (FIV) and FFV.8,272 A case of human co-infections by HIV-1 and SFV from mandrills has also been reported, without clinical consequences that could be attributed to the SFV infection.233
Role of Antiviral Restriction Factors
APOBEC3
FV genomes are sensitive to editing by a variety of cellular catalytic polypeptide 3 apolipoprotein B messenger RNA (mRNA)-editing (APOBEC) proteins.41,141,205 To edit complementary DNA (cDNA) during reverse transcription, APOBEC3 proteins have to be encapsidated into the nascent virus particle. FVs encode the accessory Bet protein to preserve genome stability. The HIV-1 Vif protein prevents APOBEC3 particle incorporation by routing it to the proteasomal degradation pathway. In contrast, Bet prevents APOBEC3 encapsidation by binding and quantitatively trapping the deaminase.36,175 The PFV Bet function was found to be broadly active against various primate APOBEC3 proteins.175 However, some species specificity was also observed, as PFV Bet was found to be inactive against all or some mouse, feline, and rhesus monkey APOBEC3 proteins, as well as against human APOBEC3DE and APOBEC3H.175 Because reverse transcription of FV RNA takes place to a significant degree in virus-producing cells (see later discussion), APOBEC3 restriction of FVs occurs at a different point in the replication cycle than that reported for orthoretroviruses.141
TRIM5α
The tripartite interaction motif (TRIM) proteins comprise a large family of cellular proteins that are components of the innate immune defense mechanism and are active at various stages of replication against many viruses, including retroviruses.167,244
TRIM5α has been studied extensively for its activity against lentiviruses (see Chapters 8 and 49) and was found to be active against some orthoretroviruses during the very early phase of infection, prior to reverse transcription, by inducing premature disassembly of viral capsids. Lentiviral gag genes have evolved in such a way that their products are not neutralized by the homologous TRIM5α but often by proteins from other species. TRIM5α proteins have a modular organization consisting of so-called N-terminal RING domains, followed by zinc-binding B boxes, coiled-coil domains, and the C-terminal variable region B30.2/SPRY.167,244 The ability to recognize and bind in a species-specific manner to retroviral capsids is mediated by the B30.2 domain.244 FV gag genes are highly divergent from their orthoretroviral counterparts; thus, it was uncertain whether TRIM5α would react against FVs and, if so, what the molecular basis would be. Nonetheless, it has been shown that primate TRIM5α proteins restrict FVs in a species-specific manner.170,263 The specificity of TRIM5α has been mapped to variable residues of the B30.2 domain, which are important for neutralization of lentiviruses, and to the N-terminal half of the FV Gag.263 The activity of TRIM5α against divergent capsid proteins, including those of FVs, which do not mature into the canonical orthoretrovial matrix (MA), capsid (CA), and nucleocapsid (NC) subunits, implies an even wider structural recognition pattern than previously assumed.
Tetherin (CD317)
CD317/tetherin is an integral membrane protein with an N-terminal membrane-spanning domain and a C-terminal glycosyl-phosphatidylinositol anchor. CD317 interacts directly with the actin cytoskeleton and blocks the release of enveloped viruses from infected cells. In HIV-1–infected cells, it is antagonized by the accessory gene product Vpu, for HIV-2 by Env, and in some SIVs by the Nef protein (see Chapters 49 and 51). The fact that different lentiviruses developed effective strategies to neutralize CD317 argues for its in vivo importance. As is the case for other enveloped viruses, CD317 is also active against FVs.104,262 A FV protein antagonizing tetherin has not yet been identified. The activity of tetherin against PFV shows some mechanistic differences in comparison to HIV-1, because dimerization-deficient tetherin inhibits PFV replication with the same efficiency as the wild-type factor.262
Interferon
All three aforementioned restriction factors are inducible by interferon (IFN) to which FVs are vulnerable.56,150 Early studies indicated already that FVs are vulnerable to IFNs,194,195 and
it has been shown that toll-like receptor 7 (TLR7) expression in plasmocytoid dendritic cells is the likely factor in sensing FV RNA resulting in the induction of type I IFN.204 Replicating virus was found to be not required for this type of IFN induction. The addition of type II IFN abolishes FV replication in vitro almost completely, whereas the activity of type I IFN is less pronounced.56,150 The type I IFN–induced protein IFP35 has been demonstrated to down-regulate BFV transcription and replication by interacting with the trans-activator protein of BFV (TasBFV).236 Furthermore, the analysis of specific PFV gag arginine to lysine conversion mutants (see later discussion) revealed a likely antiviral role of IFNs that cannot be attributed solely to the three restriction factors discussed earlier.150 Whether the IFN-inducible antiviral promyelocytic leukemia (PML) proteins play a role in restricting FV replication183 needs further substantiation; PML proteins appear not to be involved in an establishing viral latency.156
it has been shown that toll-like receptor 7 (TLR7) expression in plasmocytoid dendritic cells is the likely factor in sensing FV RNA resulting in the induction of type I IFN.204 Replicating virus was found to be not required for this type of IFN induction. The addition of type II IFN abolishes FV replication in vitro almost completely, whereas the activity of type I IFN is less pronounced.56,150 The type I IFN–induced protein IFP35 has been demonstrated to down-regulate BFV transcription and replication by interacting with the trans-activator protein of BFV (TasBFV).236 Furthermore, the analysis of specific PFV gag arginine to lysine conversion mutants (see later discussion) revealed a likely antiviral role of IFNs that cannot be attributed solely to the three restriction factors discussed earlier.150 Whether the IFN-inducible antiviral promyelocytic leukemia (PML) proteins play a role in restricting FV replication183 needs further substantiation; PML proteins appear not to be involved in an establishing viral latency.156
Virion Structure and Virion Nucleic Acid
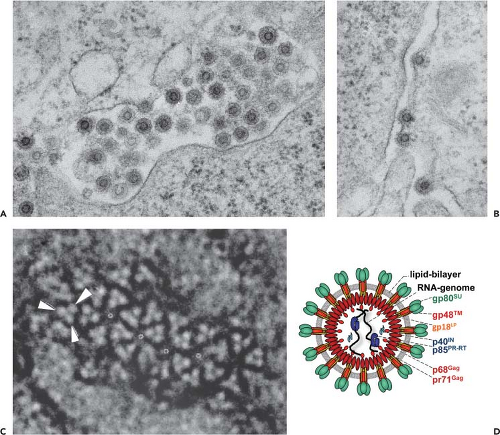
By ultrastructural analysis, FVs appear as immature-looking core particles surrounded by a lipid bilayer with embedded prominent Env proteins94,154,186,256 (Fig. 52.4). In negative-staining electron microscopy (EM), the virion has a diameter
of approximately 110 nm and a core of approximately 60 nm.94,154,186,256 The core has an immature morphology owing to the very limited cleavage of the Gag precursor protein by the viral protease (PR) (see later discussion). The cores of infectious PFV virions are made up of the Gag precursor pr71Gag and its larger processing product p68Gag at a ratio of 1:1 up to 1:4.33 This Gag doublet is seen with all FVs,71 although the nonprimate FV capsid proteins are considerably smaller than their primate relatives.92,119,199,242 The core is probably oriented radially with the Gag N-terminus pointing outward in the direction of the Env proteins256 (Fig. 52.4C). The prominent surface spikes average 15 nm in length and are organized as trimers in ring-like structures.256 A feature that PFV shares with HBV is the formation of subviral particles (SVPs) consisting only of membranous Env-containing vesicles and devoid of cores.224,228
of approximately 110 nm and a core of approximately 60 nm.94,154,186,256 The core has an immature morphology owing to the very limited cleavage of the Gag precursor protein by the viral protease (PR) (see later discussion). The cores of infectious PFV virions are made up of the Gag precursor pr71Gag and its larger processing product p68Gag at a ratio of 1:1 up to 1:4.33 This Gag doublet is seen with all FVs,71 although the nonprimate FV capsid proteins are considerably smaller than their primate relatives.92,119,199,242 The core is probably oriented radially with the Gag N-terminus pointing outward in the direction of the Env proteins256 (Fig. 52.4C). The prominent surface spikes average 15 nm in length and are organized as trimers in ring-like structures.256 A feature that PFV shares with HBV is the formation of subviral particles (SVPs) consisting only of membranous Env-containing vesicles and devoid of cores.224,228
It appears that FV particles contain fewer Pol molecules than orthoretroviruses, which is consistent with the high processivity activity of its RT.23,197 The interpretation of older studies demonstrating equal amounts of Pol in foamy and orthoretroviruses33 have been complicated by the detection of significant amounts of extraparticular Pol protein present in FV particle preparations of different origin as reported by Swiersy et al.231 This study also revealed that in sharp contrast to orthoretroviruses, FV replication tolerates a great imbalance in the relative ratios of Gag and Pol molecules in virus producing cells.231 This is owing to the unusual Pol encapsidation strategy of FVs (see later discussion), in which viral RNA is the limiting factor at conditions of high cellular levels of Pol.
The physical stability of FVs has not been directly compared with that of orthoretroviruses. However, owing to their immature core and a particular Env topology (see later discussion), FVs are probably quite stable. This is illustrated by the fact that vector particles can be concentrated more than 100-fold (i.e., by ultracentrifugation) without great loss of infectivity.65,89
Viral nucleic acid is present within the core of extracellular FV particles, and it appears to be a mixture of RNA and DNA. RNA dominates at a ratio of approximately 1:1 to 7:1, depending on the genomic region analyzed (long terminal repeat [LTR] vs. gag).203,269,270 Consistent with this, both forms of nucleic acid were detected in plasma and saliva of an experimentally infected cynomolgus macaque by PCR and RT-PCR.25 Full-length DNA was shown to be present in roughly 5% to 20% of FV virions, and functional studies using the RT-inhibiting drug AZT indicated DNA to be the relevant genome for infection, at least at the high multiplicity of infection (MOI) studied.150,160 Because DNA was found in PFV and FFV virions, the idea that FVs are facultative DNA viruses may be generalized.203
However, it has not been investigated in much detail to what extent reverse transcription is taking place late in the replication cycle. For instance, it is likely that there are genomic regions, such as the gap in the plus strand cDNA (see later discussion), that still remain single stranded. The finding of more LTR than gag region reverse transcripts in virions is a clear indication of this.203,269 In addition, because more RNA than DNA is found in virions, it would be interesting to know whether purely RNA-containing viruses exist or whether most viruses contain both forms of nucleic acids.
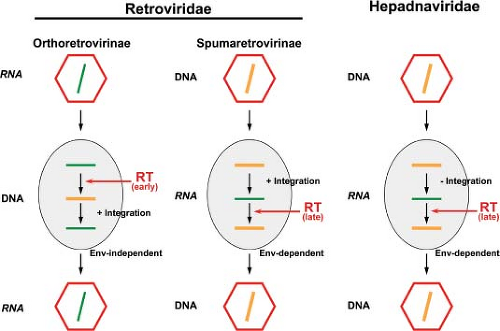
It is with respect to reverse transcription taking place at a late step in viral replication that FVs diverge at most from orthoretroviruses, which display a RNA to DNA ratio of approximately 105:1 in extracellular virions.203 The most compelling evidence for this are experiments in which the infectivity of virion-extracted FV DNA has been demonstrated.150,203,269 Thus, FVs functionally bridge the replication pathways of orthoretroviruses and hepadnaviruses (Fig. 52.5). It is because of this analogy to the hepadnaviral replication cycle (see Chapter 68) that the full-length FV RNA has been termed (pre-) genomic.
In addition to reverse transcription in late phases of FV particle morphogenesis, it has been shown that, similar to orthoretroviruses, reverse transcription also occurs during the early phases of FV replication upon target cell entry.42,270 Given
the aforementioned ratios of RNA to DNA in extracellular viruses, reverse transcription during the early phase of FV replication may only become relevant at a very low MOI, when the amount of virion DNA may be too low to sustain a productive infection.270 Furthermore, the discrepant results reported about the importance of the differentially timed reverse transcription events for FV infectivity may reflect the inherent differences in the cell types used for virus production.42,150,160,203,269,270 In essence, the generally accepted view that virions contain either an RNA or DNA genome, but not both, may not apply to FVs.
the aforementioned ratios of RNA to DNA in extracellular viruses, reverse transcription during the early phase of FV replication may only become relevant at a very low MOI, when the amount of virion DNA may be too low to sustain a productive infection.270 Furthermore, the discrepant results reported about the importance of the differentially timed reverse transcription events for FV infectivity may reflect the inherent differences in the cell types used for virus production.42,150,160,203,269,270 In essence, the generally accepted view that virions contain either an RNA or DNA genome, but not both, may not apply to FVs.
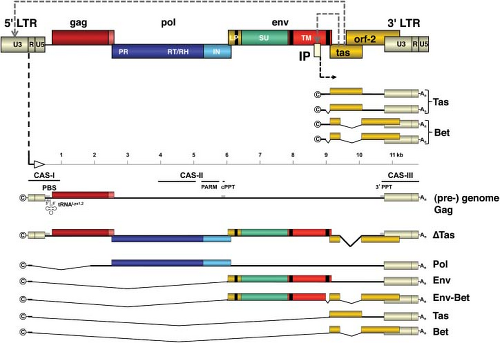
Genome Organization
The schematic representation of the genome of PFV is shown in Figure 52.6. All FV genomes share common genome structures.191 Between the LTRs, the canonical gag, pol, and env genes are found; downstream of env, the accessory open reading frames (ORFs) are found. Proviruses are 12 to 13 kb—long in comparison to those of other retroviruses. The large size of FVs genomes is partially attributed to the extraordinary long U3 regions of the LTR, which can be explained in part by the overlapping ORF-2 (see Fig. 52.6). However, the accessory ORF-2 reaches only to some extent (approximately 300 bp, in the case of PFV) into the more than 1.4 kb long U3 region, leaving several hundred bases without known function. In the latter, very few enhancer elements, such as those for AP-1 and Ets-1,152,213 and the short sequence motifs responsive to the viral transactivator (see later discussion), are present.
Stay updated, free articles. Join our Telegram channel

Full access? Get Clinical Tree