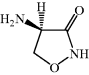
- D-Cycloserine (DCS) is a natural antibiotic with good oral bioavailability and activity against both Gram positive and Gram negative bacteria.
- D-Cycloserine interferes with two enzymes in cell-wall biosynthesis: L-alanine racemase, which forms D-alanine from L-alanine, and D-ala-D-ala-ligase, which forms the D-ala-D-ala dipeptide that is required for peptidoglycan crosslinking.
- DCS is a reserve drug for the treatment of multiple-drug-resistant tuberculosis (MDR-TB).
D-Cycloserine (DCS) (Figure 5.3.1) is an orally available antibiotic, which is used as part of a combination therapy regimen in a second-line treatment for Mycobacterium tuberculosis.
5.3.1 Discovery
DCS, which was originally referred to as oxamycin or seromycin, or by its IUPAC name (D-4-amino-3-isoxazolidone) (Kuehl et al., 1955), is a metabolite produced by Streptomyces garyphalus, orchidaceus, and lavendulae. It was originally isolated from an extract from Streptomyces lavendulae and was found to be a bactericidal agent with a broad spectrum of antibacterial activity.
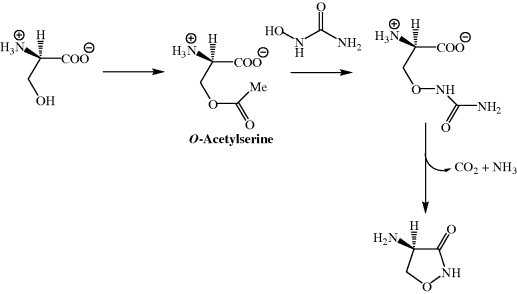
Studies in S. garyphalus have shown that the biosynthesis of DCS involves the nucleophilic attack of hydroxyurea on O-acetylserine (Scheme 5.3.1) (Svensson and Gatenbeck, 1982).
5.3.2 Synthesis
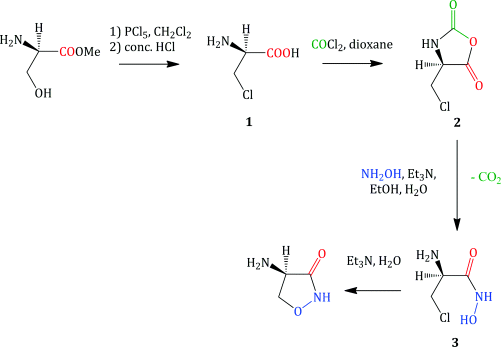
As it is a small-molecule antibiotic, DCS can be obtained by either fermentation (Shull et al., 1956; Harned, 1957) or a relatively straightforward synthesis (Scheme 5.3.2) (Plattner et al., 1957).
The original synthesis of DCS led to a racemic mixture of the cycloserine enantiomers, which had to be resolved using tartaric acid (Stammer et al., 1955). The first stereospecific DCS synthesis used as the starting material an ester of D-serine (the unnatural stereoisomer) and gave DCS in an overall yield of approx 8% (Scheme 5.3.2). The simple chemical transformations involved in this synthesis include:
- Side-chain chlorination of the serine ester (an example of a nucleophilic substitution), followed by ester hydrolysis (Shiraiwa et al., 1996) to give the β-chloro-D-alanine 1.
- Anhydride 2 formation by the reaction of the α-amino acid 1 with phosgene (COCl2).
- Formation of a hydroxamic acid 3, from the reaction of the anhydride 2 with hydroxylamine.
- Cyclisation to give DCS, with the stereochemistry at the 4-position being derived from that of the α-carbon atom of the D-serine starting material (Plattner et al., 1957).
5.3.3 Bioavailability
Due to its high water solubility and low molecular weight, DCS has an apparent volume of distribution which is similar to that of total body water (Zhu et al., 2001). Following a single oral 500 mg dose, the peak plasma concentration (Cmax) was 12–30 μg/mL, with a corresponding Tmax (time to peak plasma concentration) of 0.25–2.5 hours. The half-life can vary widely, between 4 and 30 hours, with a mean value of 10 hours. Typical MICs for DCS are approximately 10 μg/mL (Berning and Peloquin, 1999). Steady-state concentrations after a 500 mg dose of DCS every 12 hours would produce Cmax : MIC ratios of about 2.4 and 24-hour concentration : MIC ratios of approximately 1, so that serum concentrations would equal, or exceed, the MIC for most of the dosing interval. A 500 mg dose of DCS every 12 hours would thus be a suitable treatment regimen, with initial low dosages of DCS (250 mg every 12–24 hours) gradually being increased (over several days) to this level. The pharmacokinetics of DCS are minimally affected by orange juice and antacids, whereas a high-fat meal delays absorption (Zhu et al., 2001).
DCS is primarily renally excreted, so that dosage reduction is required for patients with reduced kidney function (Peloquin, 1991).
Due to its structural resemblance to D-alanine, DCS is recognised by the D-alanine proton motive-force-dependent transport system and is taken into bacterial cells by this active transport system that supplies the bacterium with amino acids from its environment. This gives DCS an enormous advantage and activity over a broad spectrum of bacteria, both Gram positive and Gram negative (Chopra, 1988).
5.3.4 Mode of Action and Selectivity
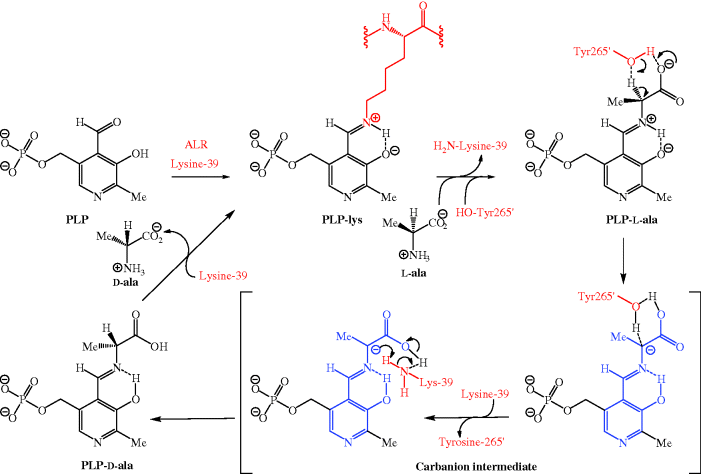
Peptidoglycan (or murein) is composed of linear glycan chains crosslinked by peptide units, and it is the nature of these peptide crosslinks which is important here, as DCS targets two enzymes that are crucial to the synthesis of the D-alanine-D-alanine (D-ala-D-ala) dipeptide unit subsequently incorporated into a pentapeptide chain. D-Ala-D-ala is required for the synthesis of the bacterial cell wall, but D-alanine is the unnatural enantiomer of alanine and, in a rare example of the destruction of chirality in nature, bacteria produce D-ala from the natural enantiomer, L-ala. Alanine racemase (ALR) catalyses this process, producing a racemic alanine mixture (50:50) starting from either alanine enantiomer, using pyridoxal 5′-phosphate (PLP) as the co-factor to cause racemisation via an imine (Schiff’s base), thereby increasing the acidity of the α-hydrogen of the alanine (Scheme 5.3.3) (Watanabe et al., 2002). Initially the PLP is bound to the enzyme through imine formation with a lysine residue (lysine-39 in Bacillus stearothermophilus) of one of the monomer units of the ALR (to give a PLP-lys complex). Trans-amination, by reaction of L-ala with the PLP-lysine, gives a new imine (PLP-L-ala) at the active site, in which the acidity of the α-hydrogen of the alanine is increased, as the positively charged N of the protonated imine is electron-withdrawing (as is the carboxyl group), and because deprotonation gives a resonance-stabilised intermediate. Deprotonation of this PLP-ala complex, by a tyrosine residue (Tyr265′ from the other half of the ALR dimer in Bacillus stearothermophilus) gives a planar carbanion intermediate (i.e. the portion of the intermediate highlighted in blue is flat, enabling the stabilisation of the intermediate through extensive delocalisation of the negative charge), which can be reprotonated from either face (i.e. from above or below), giving the PLP Schiff’s base complex of either of the pair of alanine enantiomers. A further trans-amination with the active-site lysine-39 is the final step in the catalytic cycle and regenerates the PLP-lys complex ready for reaction with another L-ala molecule (Scheme 5.3.3).
Scheme 5.3.3 Mechanism of Bacillus stearothermophilus ALR-catalysed alanine racemisation (only reprotonation from above is shown, for clarity)
Stay updated, free articles. Join our Telegram channel

Full access? Get Clinical Tree