Circoviridae
Xiang-Jin Meng
History
The prototype circovirus, porcine circovirus (PCV), was first reported in 1974 as papovavirus-like and picornavirus-like particles in a contaminated porcine kidney cell line PK-15.106 The name circovirus was proposed in 1982 when the viral genome was determined to be a circular single-stranded DNA molecule.104 PCV was not known to be pathogenic105 until 1997 when a variant strain, PCV type 2 (PCV2), was isolated from pigs with a wasting disease.2 The PK-15 cell-derived virus was designated PCV1 to distinguish it from the pathogenic PCV2. The first circoviruses identified in avian species are chicken anemia virus (CAV) and subsequently psittacine beak and feather disease virus (BFDV), both of which are associated with diseases in birds. CAV was isolated in 1979,118 although its complete genomic sequence was not determined until 1991.79 BFDV was first isolated as a novel virus with a single-stranded circular DNA genome in 1989 from cockatoos with beak and feather disease.92
Definitive evidence of human infections by known porcine or avian circoviruses is lacking. Antibodies to PCV1 were reportedly detected in humans,105 although subsequent studies could not confirm the initial report.3,28,39 Recently, novel circovirus-like DNA sequences were detected in stool samples from humans,51 although the biological and clinical significances of these novel circovirus sequences in humans remain unknown. The recent discovery of PCV1 and PCV2 DNA in live-attenuated human rotavirus vaccines prompted the U.S. Food and Drug Administration (FDA) to temporarily suspend the use of rotavirus vaccines.4,111
Classification
Viruses in the family Circoviridae infect mammalian and avian species. The genomic organization and replication strategy of circoviruses are similar to those of plant geminiviruses and nanoviruses. In fact, animal circoviruses may have evolved from a plant nanovirus through host-switch followed by a recombination event with a picorna-like virus in a mammalian host.37 Two genera of circoviruses, Circovirus and Gyrovirus, have been recognized by the International Committee on Taxonomy of Viruses (ICTV). The genus Circovirus consists of at least 11 species including BFDV, PCV1, PCV2, canary, duck, finch, goose, gull, pigeon, starling, and swan circoviruses.115 CAV, which has larger virion and genome size as well as a different genomic organization, is the sole member of the genus Gyrovirus. The Torque teno virus (TTV) and mini-TT virus, which were once classified in the Circoviridae, have now been reclassified in a new family Anelloviride.8 The “Cyclovirus” recently discovered from the stool samples of humans and chimpanzees may represent a previously unrecognized genus in the family Circoviridae.51
Virion Structure
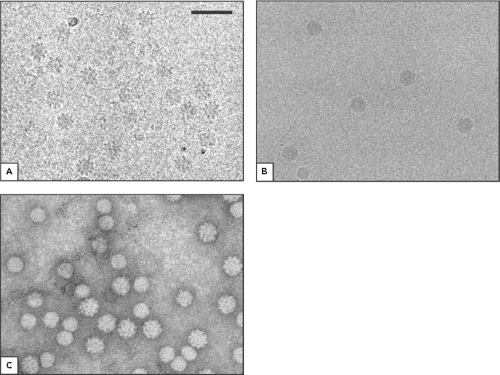
Circoviruses contain a single-stranded circular DNA genome enclosed within a capsid, which is presumably the only structural protein in the virion. Morphologically, members of the genus Circovirus appear as small, nonenveloped, icosahedral particles of approximately 15 to 20 nm in size (23,70; e-Fig. 58.1). The virion particles of CAV in the genus Gyrovirus are larger, with a diameter of 25 to 26.5 nm (23,36,64; Fig. 58.1). Members of Circoviridae all have a T=1 structure containing 60 copies of the capsid protein.23 However, the capsids of genus Circovirus consist of 12 flat pentameric morphological units, whereas the capsid of CAV in the genus Gyrovirus consists of 12 pentagonal trumpet-shaped units. Therefore, the virions of the genus Circovirus have a smoother and more featureless surface than that of CAV of the Gyrovirus (23; Fig. 58.1).
Genome Structure and Organization
The genome of Circoviridae is a single-stranded circular DNA molecule of 1.7 to 2.0 kb for genus Circovirus, and 2.3 kb for
Gyrovirus CAV.96,115 These are the smallest DNA viruses known to infect mammals and birds, and the genome size is reduced to the absolute necessities for the two basic functions of a virus: copying and packaging of viral genome.
Gyrovirus CAV.96,115 These are the smallest DNA viruses known to infect mammals and birds, and the genome size is reduced to the absolute necessities for the two basic functions of a virus: copying and packaging of viral genome.
For the genus Circovirus, the genome contains two major open-reading frames (ORFs) coding for the replicase protein (Rep) and capsid protein (Cap), respectively. The rep and cap genes are oriented in the opposite direction resulting in an ambisense genome organization. The Rep and Rep′ are produced from alternatively spliced RNA transcripts. An intergenic region between the 5′ ends of rep and cap genes contains the origin of viral genome replication (Ori), which is characterized by a stem-loop structure with a nonamer motif in its apex (e-Fig. 58.2). Three or four hexamer repeat motifs adjacent to the stem-loop serve as the binding sites for Rep and Rep′ to initiate rolling-circle replication of viral genome.33,61 The rep or Ori between PCV1 and PCV2 are fully exchangeable,5,6,7,29,30,31 indicating conserved functionality of these regions among members of the genus Circovirus.
The structure and organization of the CAV genome in the genus Gyrovirus is different from that of the genus Circovirus.22,79 The CAV genome is negative sense with approximately 2.3 kb, and contains three partially overlapping ORFs, a promoter-enhancer region, and a polyadenylation signal.74,87 The ORF1 codes for the VP1 capsid protein, and the ORF2 and ORF3 code for VP2 and VP3 nonstructural proteins. The ORF3 completely overlaps ORF2, whereas ORF2 partially overlaps ORF1. The promoter-enhancer region in the 5′ NCR of CAV genome contains four or five 21-bp direct repeats (DRs) and a 12-bp insert between the second and third DRs.65,74 Host cell transcription factors bind to the DRs and the 12-bp insert, and at least two DRs and the 12-bp insert are required for efficient transcription and replication.73,74
Stages of Replication
Among the 11 species in the genus Circovirus, only PCV1 and PCV2 can be propagated in vitro. Although the CAV of the genus Gyrovirus can be propagated in cell cultures, very little is known regarding its replication. Therefore, the knowledge of circovirus replication is derived mostly from the studies of PCV1 and PCV2, and to a lesser extent, of CAV.
Attachment, Entry, and Uncoating
Glycosaminoglycans (GAG) heparin, heparan sulfate, and chondroitin sulfate B are attachment receptors for PCV2.68 It is not surprising that PCV2 utilizes GAG as the general attachment receptors (33, e-Fig. 58.3), since the virus targets
multiple organs and tissues in infected pigs, even though cells in the monocyte and macrophage lineage are the preferential targets for PCV2 in vivo.99 A yet-to-be identified specific receptor may be needed for more efficient binding and entry of PCV2 into cells.33
multiple organs and tissues in infected pigs, even though cells in the monocyte and macrophage lineage are the preferential targets for PCV2 in vivo.99 A yet-to-be identified specific receptor may be needed for more efficient binding and entry of PCV2 into cells.33
PCV2 is internalized by dendritic cells (DCs), and the internalization was observed with both mature and immature cells including blood DCs, plasmacytoid DCs, and DC precursors, and thus suggestive of a nonmacropinocytic uptake of the virus.112,113 PCV2 virus-like particles (VLPs) quickly bind to porcine monocytic cells 3D4/31, and enter cells predominantly via clathrin-mediated endocytosis and require an acidic environment for infection.69 The epithelial cells are also major targets for PCV2 in vivo. Although PCV2 quickly attaches to epithelial cells, virus entry was slow.67 It appears that a dynamin- and cholesterol-independent, but actin- and small GTPase-dependent pathway, allows PCV2 entry and internalization leading to full replication in epithelial cells.67 After entry, PCV2 is localized in the endosomes (33, e-Fig. 58.3). As the endosomal vesicles move toward the nuclear membrane and become acidic, a serine protease appears to be required for PCV2 release from the endosome, suggesting that a proteolytic cleavage of Cap may be a part of the uncoating process.66,68,69 PCV2 infection of untreated and chloroquine diphosphate–treated PK-15 cells was blocked by a serine protease inhibitor, suggesting that serine protease–mediated PCV2 disassembly is enhanced in porcine epithelial cells but inhibited in monocytic cells after inhibition of endosome-lysosome system acidification.66
Transcription
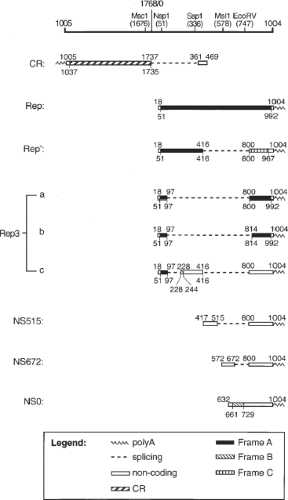
A total of nine RNA transcripts were synthesized during productive PCV2 infection in PK-15 cells: Cap protein RNA (CR), five Rep-associated RNAs (Rep, Rep′, Rep3a, Rep3b, and Rep3c), and three NS-associated RNAs (NS515, NS672, and NS0) (21; Fig. 58.2). Rep′, Rep3a, Rep3b, and Rep3c are produced from Rep by alternate splicing. The three NS-associated RNAs are transcribed from three different promoters inside ORF1, and share only the 3′ sequence with Rep.21 A stop codon introduced at the 5′-end of CR did not affect Rep-associated viral antigen or DNA synthesis.14 Altering the consensus dinucleotides at the splice junctions of the minor Rep- and NS-associated RNAs or introducing a stop codon in the abundant NS0 RNA also had no effect on viral protein or DNA synthesis. However, mutations resulting in truncated Rep or Rep´ reduced viral protein synthesis by more than 99% and abolished viral DNA replication, indicating that both Rep and Rep´ are essential for PCV2 replication.20 In contrast to the pathogenic PCV2, a total of 12 RNAs were synthesized in PCV1-infected PK15 cells14 including the viral CR RNA, eight Rep-associated RNAs, and three NS-associated RNAs.14 The promoter for cap is mapped within the ORF1 (nt 1328–1252), and the promoter for rep is located in the intergenic region (nt 640–796) and overlaps the Ori of PCV1.62
Several cellular gene transcripts were upregulated in both PCV2-infected PK-15 cells and affected tissues including two transcripts with homology to an RNA splicing factor (SPF30) and a hyaluronan-mediated motility receptor (RHAMM).10 Microarray analyses of the genes in lymph nodes of PCV2-infected pigs revealed altered expression levels in genes that are involved in innate immune defense (TLR1, CD14, and CD180), immunosuppressed responses (FGL2 and GPNMB), pro-inflammatory signals (galectin-3) and fasting processes (Angiopoietin-like 4, ANGPTL-4), suggesting that PCV2 has developed an intricate mechanism to induce immunosuppression, inflammatory cell infiltration and weight loss in pigs.49 PCV2, but not PCV1, induces interleukin 10 (IL-10) secretion by monocytic cells, which led to repression of IL-12 in peripheral blood mononuclear cells (PBMCs).45 The PCV2 ORF3 protein binds to a regulator of G protein signaling (RGS), and co-localized with poRGS16 in lipopolysaccharide (LPS)-activated porcine PBMC. The poRGS16 appeared to participate in the translocation of ORF3 protein into the nucleus.103
NF-kB was activated concomitantly with PCV2 replication, and treatment of cells with an NF-kB inhibitor reduced virus protein expression and virion production, suggesting that NF-kB activation is important for PCV2 replication.114
NF-kB was activated concomitantly with PCV2 replication, and treatment of cells with an NF-kB inhibitor reduced virus protein expression and virion production, suggesting that NF-kB activation is important for PCV2 replication.114
Three viral proteins of CAV in the genus Gyrovirus are derived from a single 2.0-kb mRNA species.76,85 Several minor mRNA species of 1.6, 1.3, and 1.2 kb in size are also identified (42, e-Fig. 58.4). The 1.3-kb RNA had a splice site joining nt 1222 to nt 1814 and encoded head-to-tail VP1. The 1.2 kb RNA possessed a splice site joining nt 994 to nt 1095 and encoded several putative proteins with frameshift mutations. CAV contains a single promoter–enhancer region with four consensus cyclic AMP response element sequences that are similar to the estrogen response element consensus half-sites. These sequences are arranged as direct repeats, an arrangement that can be recognized by members of the nuclear receptor superfamily, and may provide a mechanism to regulate CAV activity in situations of low virus copy number.65
Translation
The genome of genus Circovirus consists of two major ORFs: ORF1 encodes the Rep and Rep′, and the ORF2 encodes Cap. Rep is translated from the full-length rep transcript, whereas Rep′ is produced from a spliced transcript.33 Both Rep and Rep′ are essential for the initiation of virus replication.19,59,60 Mutation within motifs I to III and the putative dNTP-binding (GKS) box of the Rep and Rep′ interfered with viral replication. Motifs I to III are essential for PCV1 Ori cleavage.101 The repression of rep promoter is mediated by binding of Rep to H1 and H2 hexamers in the Ori of PCV1; however, transcription of cap promoter is not influenced by viral proteins.33,60 Both Rep and Rep′ co-localize in the nucleus of infected cells and form homomeric and heteromeric complexes.34,59 Three putative nuclear localization signals (NLSs) are present in the N-termini of Rep/Rep′: NLS1 and NSL2 mediate nuclear accumulation, whereas NSL3 enhances the nuclear transport of Rep and Rep′.34 PCV2 Rep interacted with an intermediate filament protein, similar to human syncoilin, and with the transcriptional regulator c-myc.102 The PCV Rep also binds three porcine cellular proteins1,35: ZNF265 is an alternative component of the spliceosome, whereas VG5Q and TDG were linked to transcriptional regulation.
The Cap of the genus Circovirus can self-assemble into VLPs,11,70,117 and it elicits neutralizing antibodies in vaccinated animals.9,97 At least five different but overlapping conformational epitopes were identified within residues 47 to 63 and 165 to 200 and the last four amino acids at the C terminus of PCV2 Cap.50,57 Two amino acid mutations in the Cap, P110A and R191S, enhance the growth ability of PCV2 in vitro but attenuate PCV2 in vivo.32 The Cap localized in the nucleoli of PCV2-infected cells54 and in the nucleoli of cells at an early stage of PCV1 infection.34 The PCV Cap interacts with numerous cellular proteins35,102 including complement factor C1qB, E3 ubiquitin ligase family member MKRN1, and proapoptotic gene product Par-4.33 The exact function of these cellular factors in PCV replication remains to be elucidated. An ORF3 has been identified in PCV1 but is truncated in PCV2. The in vitro–expressed ORF3 protein of PCV2-induced apoptosis through the activation of caspase 8 and caspase 3 pathways.52 The ORF3 protein interacts with pPirh2 and competes with p53 in binding to Pirh2 and mediates the deregulation of p53 homeostasis, leading to increased p53 levels and apoptosis of the infected cells.44,53 It was reported that abrogation of the ORF3 function attenuated PCV2 in pigs,43 although other studies showed that PCV2 pathogenicity is not solely determined by ORF3.13,41 In fact, whether or not PCV2 infection causes apoptosis remains controversial.48,58,91,113
Stay updated, free articles. Join our Telegram channel

Full access? Get Clinical Tree