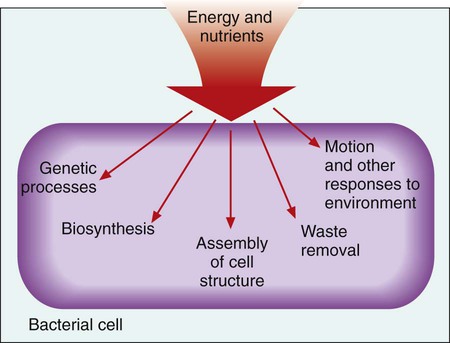
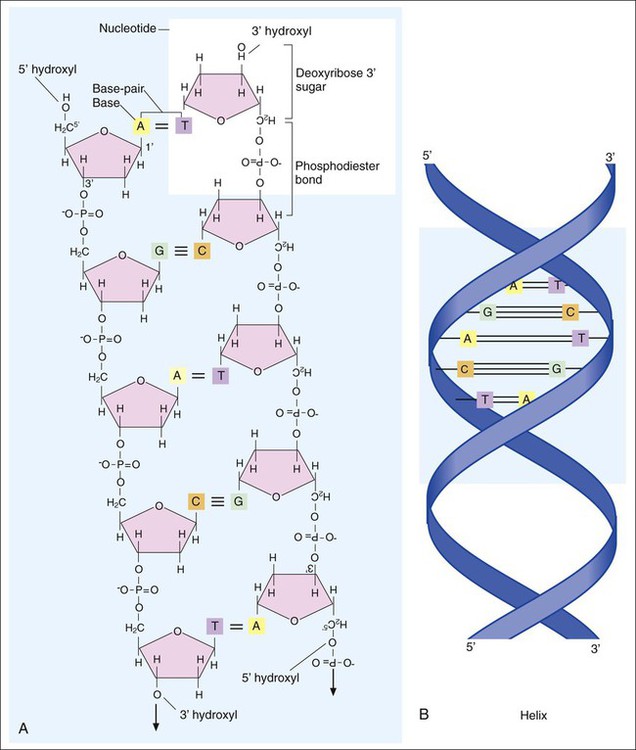
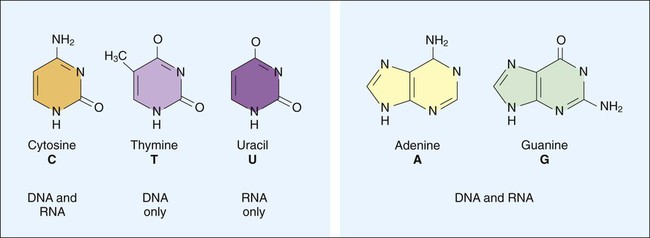
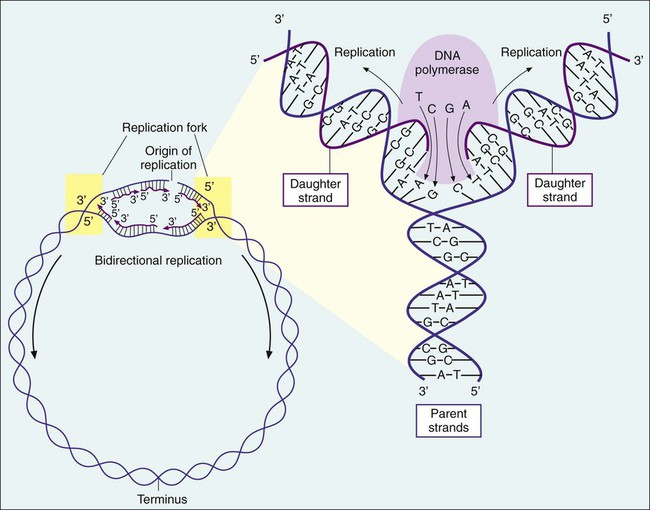
1. Describe the basic structure and organization of prokaryotic (bacterial) chromosomes, including number, relative size, and cellular location. 2. Outline the basic processes and essential components required for genetic information transfer in replication, transcription, translation, and regulatory mechanisms. 3. Define mutation, recombination, transduction, transformation, and conjugation. 4. Describe how genetic alterations and diversity provide a mechanism for evolution and survival of microorganisms. 5. Differentiate environmental oxygenation and final electron acceptors (aerobes, facultative anaerobes, and strict anaerobes) in the formation of energy. 6. Compare and contrast the key structural elements, cellular organization, and types of organisms classified as prokaryotic and eukaryotic. 7. State the functions and biologic significance of the following cellular structures: the outer membrane, cell wall, periplasmic space, cytoplasmic membrane, capsule, fimbriae, pili, flagella, nucleoid, and cytoplasm. 8. Differentiate the organization and chemical composition of the cell envelope for a gram-positive and a gram-negative bacterium. Microbial genetics, metabolism, and structure are the keys to microbial viability and survival. These processes involve numerous pathways that are widely varied, often complicated, and frequently interactive. Essentially, survival requires energy to fuel the synthesis of materials necessary to grow, propagate, and carry out all other metabolic processes (Figure 2-1). Although the goal of survival is the same for all organisms, the strategies microorganisms use to accomplish this vary substantially. • The mechanism or mechanisms by which microorganisms cause disease • Developing and implementing optimum techniques for microbial detection, cultivation, identification, and characterization • Understanding antimicrobial action and resistance • Developing and implementing tests for the detection of antimicrobial resistance • The structure and organization of genetic material • Replication and expression of genetic information • The mechanisms by which genetic information is altered and exchanged among bacteria DNA consists of deoxyribose sugars connected by phosphodiester bonds (Figure 2-2, A). The bases that are covalently linked to each deoxyribose sugar are the key to the genetic code within the DNA molecule. The four bases include two purines, adenine (A) and guanine (G), and the two pyrimidines, cytosine (C) and thymine (T) (Figure 2-3). In RNA, uracil replaces thymine. The combined sugar, phosphate, and a base form a single unit referred to as a nucleotide (adenosine triphosphate [ATP], guanine triphosphate [GTP], cytosine triphosphate [CTP], and thymine triphosphate [TTP]). DNA and RNA are nucleotide polymers (i.e., chains or strands), and the order of bases along a DNA or RNA strand is known as the base sequence. This sequence provides the information that codes for the proteins that will be synthesized by microbial cells; that is, the sequence is the genetic code. The intact DNA molecule is composed of two nucleotide polymers. Each strand has a 5’ (prime) phosphate and a 3’ (prime) hydroxyl terminus (see Figure 2-2, A). The two strands run antiparallel, with the 5’ of one strand opposed to the 3’ terminal of the other. The strands are also complementary, because the adenine base of one strand always binds to the thymine base of the other strand by means of two hydrogen bonds. Similarly, the guanine base of one strand always binds to the cytosine base of the other strand by means of three hydrogen bonds. As a result of the molecular restrictions of these base pairings, along with the conformation of the sugar-phosphate backbones oriented in antiparallel fashion, DNA has the unique structural conformation often referred to as a “twisted ladder” or double helix (see Figure 2-2, B). Additionally, the dedicated base pairs provide the format essential for consistent replication and expression of the genetic code. In contrast to DNA, which carries the genetic code, RNA rarely exists as a double-stranded molecule. The three major types of RNA (messenger RNA [mRNA], transfer RNA [tRNA], and ribosomal RNA [rRNA]) play key roles in gene expression. Certain genes are widely distributed among various organisms while others are limited to particular species. Also, the base pair sequence for individual genes may be highly conserved (i.e., show limited sequence differences among different organisms) or be widely variable. As discussed in Chapter 8, these similarities and differences in gene content and sequences are the basis for the development of molecular tests used to detect, identify, and characterize clinically relevant microorganisms. Bacteria multiply by cell division, resulting in the production of two daughter cells from one parent cell. As part of this process, the genome must be replicated so that each daughter cell receives an identical copy of functional DNA. Replication is a complex process mediated by various enzymes, such as DNA polymerase and cofactors; replication must occur quickly and accurately. For descriptive purposes, replication may be considered in four stages (Figure 2-4): 1. Unwinding or relaxation of the chromosome’s supercoiled DNA 2. Separation of the complementary strands of the parental DNA so that each may serve as a template (i.e., pattern) for synthesis of new DNA strands 3. Synthesis of the new (i.e., daughter) DNA strands 4. Termination of replication, releasing two identical chromosomes, one for each daughter cell Gene expression begins with transcription. During transcription the DNA base sequence of the gene (i.e., the genetic code) is converted into an mRNA molecule that is complementary to the gene’s DNA sequence (Figure 2-5). Usually only one of the two DNA strands (the sense strand) encodes for a functional gene product. This same strand is the template for mRNA synthesis. The next phase in gene expression, translation, involves protein synthesis. Through this process the genetic code in mRNA molecules is translated into specific amino acid sequences that are responsible for protein structure and function (see Figure 2-5). Before addressing the process of translation, a discussion of the genetic code that is originally transcribed from DNA to mRNA and then translated from mRNA to protein is warranted. The code consists of triplets of nucleotide bases, referred to as codons; each codon encodes for a specific amino acid. Because there are 64 different codons for 20 amino acids, an amino acid can be encoded by more than one codon (Table 2-1). Each codon is specific for a single amino acid. Therefore, through translation, the codon sequences in mRNA direct which amino acids are added and in what order. Translation ensures that proteins with proper structure and function are produced. Errors in the process can result in aberrant proteins that are nonfunctional, underscoring the need for translation to be well controlled and accurate. TABLE 2-1 The Genetic Code as Expressed by Triplet-Base Sequences of mRNA*
Bacterial Genetics, Metabolism, and Structure
Bacterial Genetics
Nucleic Acid Structure and Organization
Nucleotide Structure and Sequence
DNA Molecular Structure
Genes and the Genetic Code
Replication and Expression of Genetic Information
Replication
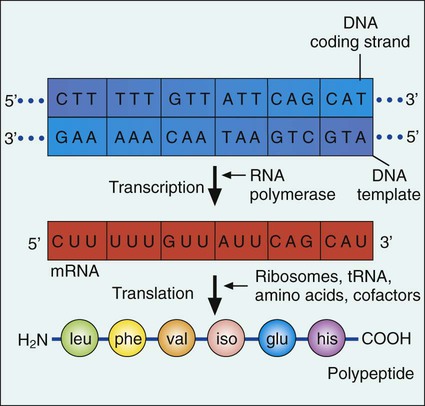
Expression of Genetic Information
Transcription.
Translation.
Codon
Amino Acid
Codon
Amino Acid
Codon
Amino Acid
Codon
Amino Acid
UUU
Phenylalanine
CUU
Leucine
GUU
Valine
AUU
Isoleucine
UUC
Phenylalanine
CUC
Leucine
GUC
Valine
AUC
Isoleucine
UUG
Leucine
CUG
Leucine
GUG
Valine
AUG (start)†
Methionine
UUA
Leucine
CUA
Leucine
GUA
Valine
AUA
Isoleucine
UCU
Serine
CCU
Proline
GCU
Alanine
ACU
Threonine
UCC
Serine
CCC
Proline
GCC
Alanine
ACC
Threonine
UCG
Serine
CCG
Proline
GCG
Alanine
ACG
Threonine
UCA
Serine
CCA
Proline
GCA
Alanine
ACA
Threonine
UGU
Cysteine
CGU
Arginine
GGU
Glycine
AGU
Serine
UGC
Cysteine
CGC
Arginine
GGC
Glycine
AGC
Serine
UGG
Tryptophan
CGG
Arginine
GGG
Glycine
AGG
Arginine
UGA
None (stop signal)
CGA
Arginine
GGA
Glycine
AGA
Arginine
UAU
Tyrosine
CAU
Histidine
GAU
Aspartic
AAU
Asparagine
UAC
Tyrosine
CAC
Histidine
GAC
Aspartic
AAC
Asparagine
UAG
None (stop signal)
CAG
Glutamine
GAG
Glutamic
AAG
Lysine
UAA
None (stop signal)
CAA
Glutamine
GAA
Glutamic
AAA
Lysine ![]()
Stay updated, free articles. Join our Telegram channel

Full access? Get Clinical Tree