Adenoviridae
Arnold J. Berk
Adenoviruses were first isolated and characterized in 1953 by two groups who were searching for the etiologic agents of acute respiratory infections.134,283 The two isolated viruses were related and named adenoviruses, after the original tissue (adenoids) in which they were discovered. Human adenoviruses are responsible for only a small portion of acute respiratory morbidity in the general population and for about 5% to 10% of respiratory illness in children. But they can be responsible for epidemic outbreaks in nursing homes and among military recruits.174 Oral administration of enteric-coated live human adenovirus (HAdV) species B types 4 and 7, the serotypes responsible for most outbreaks among military recruits, was introduced in the 1970s and was an effective vaccine for this population. But cessation of vaccine production in 1998 led to a resumption of adenovirus-associated respiratory illness at US military training centers,153,221 resulting in severe pneumonia and one death from HAdV species B type 14.330 Consequently, there is a renewed effort to produce adenovirus vaccines.166,204
More than 100 members of the adenovirus group have been identified that infect a wide range of vertebrate hosts. All of these viruses contain a linear, double-stranded DNA (dsDNA) genome encapsidated in an icosahedral protein shell with fiber proteins of varying lengths extending from the vertices of the icosahedron that bind to receptors on host-cell surfaces through a terminal globular domain.
In humans, besides respiratory disease, adenoviruses cause conjunctivitis242 and infantile gastroenteritis.9 In immunocompetent patients, adenoviruses usually cause a mild, self-limiting acute infection. However, in neonates and immunosuppressed patients, including patients with AIDS, adenoviruses can cause fulminant fatal pneumonia, hepatitis, and/or encephalitis.192,331 Laboratory diagnosis is most rapidly done by polymerase chain reaction (PCR)-based assays.161,191
In 1962, Trentin and colleagues342 made a seminal discovery: human adenovirus type 12 induces malignant tumors after inoculation into newborn hamsters. This was the first time that a human virus was discovered to be oncogenic. No epidemiologic evidence has been reported linking adenoviruses with malignant disease in humans; extensive searches have generally failed to find adenovirus nucleic acids in human tumors.208 While in one recent study HAdV DNA was detected in >50% of pediatric central nervous system (CNS) tumors by PCR, it was also detected in normal CNS tissue from the same patients.171 Thus, the CNS may be a common site of latent, persistent adenovirus infection in children. While there is as yet little evidence that adenoviruses contribute to the etiology of human tumors, the ability to induce tumors in animals and to transform cultured primary cells established adenovirus as an important model system for probing the mechanisms of oncogenesis.
As the interest in adenoviruses as tumor viruses intensified, their virtues as an experimental system became evident. The prototype human adenoviruses are easily propagated to produce high-titer stocks, and they initiate synchronous infections of established cell lines. Further, the viral genome is readily manipulated, facilitating the study of adenovirus gene functions by directed mutational analysis. Studies of adenovirus-infected cells have made numerous contributions to our understanding of viral and cellular gene expression and regulation, DNA replication, cell-cycle control, and cellular growth regulation. Perhaps the most recognized contribution of the adenovirus system to modern biology was the discovery of messenger RNA (mRNA) splicing.24,53 Today, the utility of adenovirus as a vector for gene therapy is the subject of intense exploration. This chapter overviews the structure of the adenovirus particle, the adenovirus replication cycle in human cells, and its interactions with host cells and host organisms.
Classification
Adenoviridae have been isolated only from vertebrates—however from every major class from fish to mammals. Bioinformatic analysis of genome sequences71 indicate four major lineages corresponding to four genera: Mastadenovirus, isolated from mammals, including all human adenoviruses; Aviadenovirus, isolated from birds; Atadenovirus, so named because of their unusually high A+ T content, isolated from reptiles, birds, a marsupial and mammals; and Siadenovirus, isolated from a reptile and birds. Fifty-six human adenovirus types have been recognized and are classified into seven species (A–G) on the basis of
serology, hemagglutination, oncogenicity in rodents, transformation of cultured primary cells, and genome sequencing152,280 (Table 55.1). A proposal has been made recently for systematically naming human adenovirus types (superseding the term “serotype”) based on genome DNA sequences.12 Types are to be defined by the amino acid sequence of the major capsid protein hexon, which contains the major neutralizing epitopes. These same authors have proposed that intertypic human adenovirus recombinants be designated by the identity of the hexon gene (H) and that of the fiber gene (F) encoding the second most important neutralizing epitope. For example, HAdV-H7/F3 will designate a virus with a type 7 hexon and a type 3 fiber.12 This parsimonious nomenclature makes it unnecessary to assign a new adenovirus type number to each new recombinant between previously established types that have been isolated from patients apparently infected simultaneously with two or more types in military recruiting centers and nursing homes.
serology, hemagglutination, oncogenicity in rodents, transformation of cultured primary cells, and genome sequencing152,280 (Table 55.1). A proposal has been made recently for systematically naming human adenovirus types (superseding the term “serotype”) based on genome DNA sequences.12 Types are to be defined by the amino acid sequence of the major capsid protein hexon, which contains the major neutralizing epitopes. These same authors have proposed that intertypic human adenovirus recombinants be designated by the identity of the hexon gene (H) and that of the fiber gene (F) encoding the second most important neutralizing epitope. For example, HAdV-H7/F3 will designate a virus with a type 7 hexon and a type 3 fiber.12 This parsimonious nomenclature makes it unnecessary to assign a new adenovirus type number to each new recombinant between previously established types that have been isolated from patients apparently infected simultaneously with two or more types in military recruiting centers and nursing homes.
Table 55.1 Classification Schemes for Human Adenoviruses (HAdVs, Genus Mastadenovirus) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Virion Structure
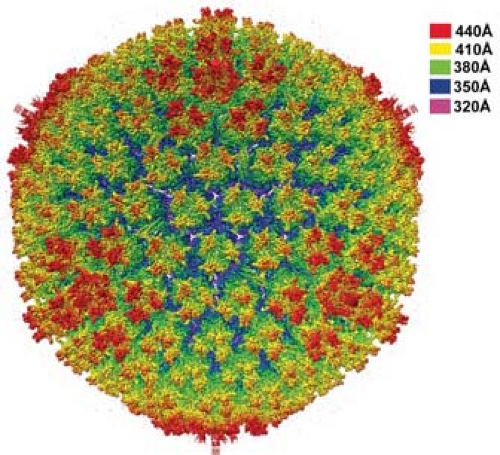
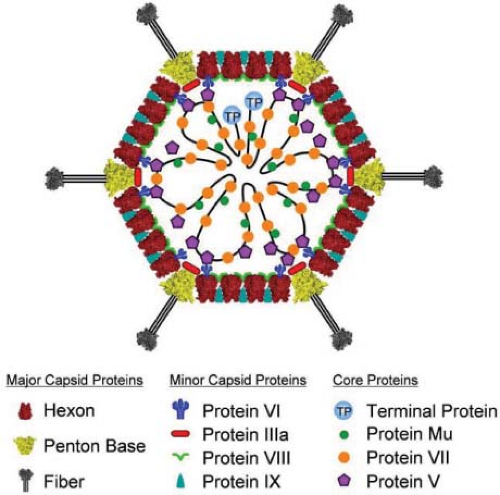
Adenoviruses are nonenveloped icosahedral particles ∼90 nm in diameter (Fig. 55.1) with fibers projecting from the vertices of the icosahedron (Fig. 55.2). Most structural studies of adenoviruses have focused on the closely related human adenoviruses types 2 and 5. The particles (virions) have a mass of ∼150 × 106 D351 and contain DNA (13% of mass), protein (87% of mass), no membrane or lipid, and trace amounts of carbohydrate because the virion fiber protein is modified by addition of glucosamine.148 Virions consist of a protein shell (capsid) surrounding a DNA-containing core. Virion polypeptides were initially characterized by disruption of isolated virions with sodium dodecylsulfate and gel electrophoresis.209,351 Comparison of electrophoretic results with genomic open reading frames (ORFs) suggests there are 12 virion proteins numbered by convention II-IX, IIIa, IVa, m, terminal protein, and the p23 viral protease, with no polypeptide I because the moiety originally designated I proved to be a mixture of aggregated molecules (Fig. 55.3, Table 55.2).
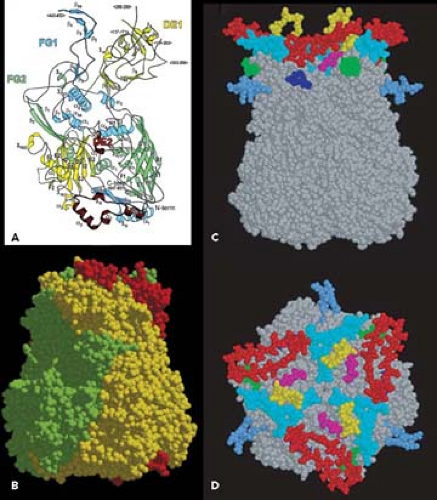
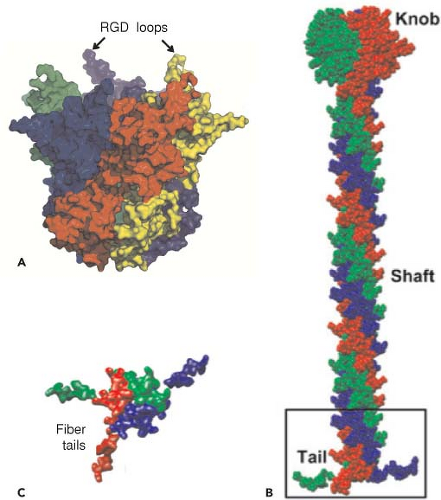
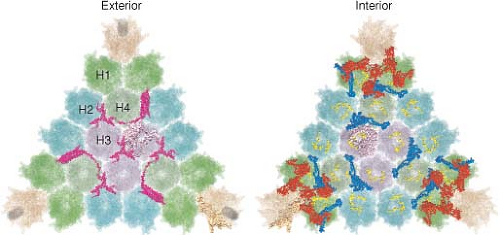
The current highest resolution structure of the Ad5 virion is based on 3.6 Å resolution cryo-electron microscopic (cryoEM)194 and 3.5 Å resolution x-ray crystallographic274 structures of the entire virion (Fig. 55.1), an ∼3 Å resolution x-ray crystal structure of the isolated hexon trimer285,286 (Fig. 55.4), and ∼1.5 Å resolution structures of the penton base403 (Fig. 55.5A), fiber shaft,353 and fiber knob352 (Fig. 55.5B). The icosahedral shell is composed primarily of 240 capsomeres of hexon trimers (12 per triangular facet of the icosahedron), 12 pentameric penton capsomeres at each vertex of the icosahedron, and 12 fibers extending from the pentons, each a trimer of the fiber polypeptide. Loops with hypervariable sequence on the external surface of hexons are important for type-specific immunogenicity and neutralization286 (Figs. 55.4C and 55.4D).
Only the bases of the flexible fibers were visualized in the cryoEM structure that depends on methods that average density signals from multiple virions. However, the full lengths of fibers are evident in negatively stained transmission electron micrographs of single virions (Fig. 55.2). Most human adenoviruses encode a single type of fiber. But types 40, 41,162 and 52152 encode two fiber proteins, with one or the other bound to each penton base. In contrast, avian adenoviruses also encode two fiber proteins, but one of each binds to each penton base.132 Because the fiber knob (Fig. 55.5B) interacts with a cellular receptor protein, the incorporation of two fiber proteins
might extend the range of cell types to which these viruses bind. The fiber shaft of the HAdVs is composed of repeats of an ∼15-residue structural motif,353 and the length of the shaft varies among types from six repeating units in Ad3 to 21 in Ad2 and Ad5. An unusual “symmetry mismatch” occurs in the interaction between the fivefold symmetric penton base and the threefold symmetric trimeric fiber. The interaction is mediated by a hydrophobic ring around a central pore on the top surface of the penton base, hydrophobic residues on the bottom of the fiber shaft, and flexible N-terminal tails (aa 10–19) of the fiber monomers (Figs. 55.5B and 55.5C) that insert into three of five available grooves formed by neighboring subunits of the penton base.196,403 The N-terminal residues of the fiber monomers (aa 1–9) extend to the bases of penton loops196 containing RGD sequences that bind to integrins on the target cell plasma
membrane, triggering endocytosis of the virion (see Mechanism of Entry section).
might extend the range of cell types to which these viruses bind. The fiber shaft of the HAdVs is composed of repeats of an ∼15-residue structural motif,353 and the length of the shaft varies among types from six repeating units in Ad3 to 21 in Ad2 and Ad5. An unusual “symmetry mismatch” occurs in the interaction between the fivefold symmetric penton base and the threefold symmetric trimeric fiber. The interaction is mediated by a hydrophobic ring around a central pore on the top surface of the penton base, hydrophobic residues on the bottom of the fiber shaft, and flexible N-terminal tails (aa 10–19) of the fiber monomers (Figs. 55.5B and 55.5C) that insert into three of five available grooves formed by neighboring subunits of the penton base.196,403 The N-terminal residues of the fiber monomers (aa 1–9) extend to the bases of penton loops196 containing RGD sequences that bind to integrins on the target cell plasma
membrane, triggering endocytosis of the virion (see Mechanism of Entry section).
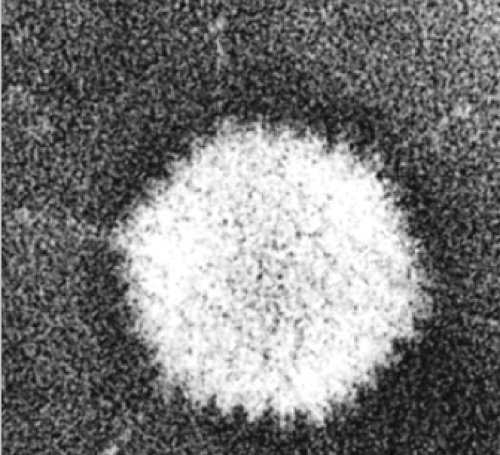 Figure 55.2. Visualization of adenovirus fibers in a negatively stained transmission electron micrograph. (Courtesy of Robley C. Williams.) |
Table 55.2 Adenovirus 5 Structural Proteins | |||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| |||||||||||||||||||||||||||||||||||||||
Minor capsid proteins IIIa, VIII, and IX stabilize nonequivalent interactions between hexon trimers, allowing the same hexon trimer to be used in four different chemical environments on the surface of the capsid194,274 (Figs. 55.6 and 55.7). Networks of interactions between the minor capsid proteins stabilize two groups of capsomeres and hold them together: a group-of-nine hexon trimers on each triangular facet of the icosahedron,268 and a group-of-six capsomeres at each vertex composed of five hexon trimers with a central penton-base pentamer (Fig. 55.7).
The core of the virion contains seven known viral proteins and the viral genome. Polypeptides VII (174 amino acids), the major core protein with over 800 copies per virion,351 and polypeptides V (368 amino acids) and m (36 amino acids) are basic, arginine-rich proteins141,284 that contact the viral DNA8,45 and likely condense the viral DNA within the core. Protein VI associates with a cavity on the inner surface of hexon trimers261,274,287 (Fig. 55.7B) and with protein V,217 tethering the highly ordered capsid to the less ordered DNA-protein core. A basic 115-residue disordered region in the middle of each polypeptide VI is proposed to contribute to an interaction with the core.288 The N-terminus of each penton-base monomer in the pentamer also interacts with the core, and the C-terminus of protein IIIa may as well,194 consistent with reports that these proteins also interact with protein V33,87,300 and that IIIa is responsible for type-specific packaging of viral DNA into virions.206 Protein IVa2 is present in only a few copies at one vertex of the icosahedron,55 binds to the packaging sequence at the left end of the genome, and is required for packaging viral DNA into the capsid (see Virion Assembly). The sixth protein in the core is the terminal protein, which is covalently attached to the 5′ ends of the viral DNA and therefore present in only two copies per virion. As discussed in Viral DNA Replication below, the terminal protein serves as a primer for DNA replication. The core also contains about 10 molecules of the p23 viral cysteine protease that functions to cleave precursors of several virion proteins during assembly and maturation of the virus particle and in virus disassembly and escape from endosomes during the infection process.367
Genome Structure and Organization
Adenovirus genomes are linear dsDNAs ranging in size from 26 to 45 kb.71 A protein primer of DNA replication called terminal protein is covalently bound to the 5′ phosphate of each strand (see Viral DNA Replication). Homologous genes shared by all adenoviruses encode the three viral proteins required for viral DNA replication, the terminal protein, viral DNA polymerase, and viral single-stranded DNA (ssDNA) binding protein, and major structural components of the virion described (see Virion Structure), except that genes encoding the core protein V are found only in the mastadenoviruses isolated from mammals.71 These common genes are encoded in the central portion of the genome. Additional genes specific to adenoviruses from different vertebrates are encoded primarily at the termini of the genome. Also, the genomes of all adenoviruses have inverted terminal repeat sequences ranging in size from 36 to over 200 bp.71 As discussed in Viral DNA Replication below, the inverted repeats function as DNA replication origins at each end of the viral genome and enable single strands of viral DNA displaced during asymmetric strand synthesis to circularize by base-pairing of their terminal sequences. The resulting
base-paired “panhandles” in the otherwise ssDNA molecules function as origins for replication of displaced single strands.
base-paired “panhandles” in the otherwise ssDNA molecules function as origins for replication of displaced single strands.
Most human adenovirus genomes encode homologous genes represented by the extensively studied, closely related human species C adenoviruses types 2 and 5 (Fig. 55.8). HAd2 DNA was the first adenovirus genome to be completely sequenced,279 and it includes a total of 35,937 base pairs (bp). This chapter will concentrate on the human adenoviruses, all having the same general genome organization.
The human adenovirus genomes include several repeats of a cis-acting packaging sequence between the left terminal repeat and the first protein-coding region (E1A).129 This packaging sequence must be located within several hundred bp of an end of the chromosome to direct the proper packaging of viral DNA into infectious virions.129
The human adenovirus genomes contain five early transcription units (E1A, E1B, E2, E3, and E4), four intermediate transcription units transcribed at the onset of viral DNA replication (IX, IVa2, L4 intermediate, and E2 late), and one late transcription unit (major late) that is processed to generate five families of late mRNAs (L1–L5), all of which are transcribed by RNA polymerase II (Fig. 55.8). As discussed in
the Late Transcription section, the major late promoter is also transcribed at a low level early during infection, generating one of the L1 family mRNAs encoding the p52/55 kD protein (a 48-kD polypeptide that runs as an apparent doublet of 52 and 55 kD on SDS polyacrylamide gels because of variation in the degree of phosphorylation). Also, after the onset of viral DNA replication, the E2 transcription unit is transcribed from an alternative promoter called the E2 late promoter as well as the early E2 promoter active before and after the onset of viral DNA replication. The chromosome also carries one or two (depending on the type) virus-associated (VA) genes transcribed by RNA polymerase III. By convention, the map is drawn with the E1A gene at the left end. Both strands of the viral DNA are transcribed with the rightward reading strand as the template for the E1A, E1B, IX, major late, VA RNA, and E3 transcription units, and the leftward reading strand as the template for the E4, E2, and IVa2 units.
the Late Transcription section, the major late promoter is also transcribed at a low level early during infection, generating one of the L1 family mRNAs encoding the p52/55 kD protein (a 48-kD polypeptide that runs as an apparent doublet of 52 and 55 kD on SDS polyacrylamide gels because of variation in the degree of phosphorylation). Also, after the onset of viral DNA replication, the E2 transcription unit is transcribed from an alternative promoter called the E2 late promoter as well as the early E2 promoter active before and after the onset of viral DNA replication. The chromosome also carries one or two (depending on the type) virus-associated (VA) genes transcribed by RNA polymerase III. By convention, the map is drawn with the E1A gene at the left end. Both strands of the viral DNA are transcribed with the rightward reading strand as the template for the E1A, E1B, IX, major late, VA RNA, and E3 transcription units, and the leftward reading strand as the template for the E4, E2, and IVa2 units.
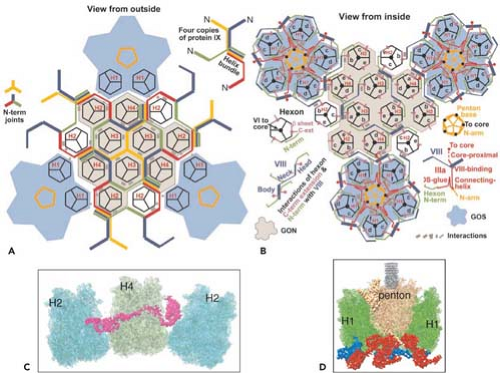 Figure 55.7. Diagram of the interactions of minor proteins IIIa, VIII, and IX that stabilize nonequivalent hexon trimer interactions. A: Hexon trimers are labeled H1 to H4 as described in Figure 55.6. The diagram at the upper right represents four different conformations of protein IX shown in different colors. N indicates their amino termini. These link hexon trimers in a group of nine (gray), and link groups of nine from neighboring facets. Groups-of-nine hexon trimers are released when virions are disrupted under mild conditions.268 B: Letters a through f indicate hexon monomers with distinct conformations of their extended N- and C-termini. As diagrammed at the lower left, three hexon C-termini and four hexon N-termini interact with one protein VIII. At each vertex of the icosahedron, five copies of protein IIIa link five hexon trimers to the central penton base, forming a group-of-six capsomeres (GOS, shaded in blue). Protein VIII links groups-of-nine hexon trimers (GON) to groups-of-nine hexon trimers in neighboring facets and groups-of-nine hexon trimers to groups-of-six capsomeres at the vertices. Protein VI binds in a central cavity on the inner surface of hexon trimers, represented as a black circle. C: Side view of protein IX (purple) inlaid into the canyons between hexon trimers. D: Side view of proteins IIIa (red) interacting with penton base and H1 hexons and protein VIII (blue) interacting with H1 hexons on the inner surface of the capsid shell. (Courtesy of Z. Hong Zhou. Adapted from Liu H, Jin L, Koh SB, et al. Atomic structure of human adenovirus by cryo-EM reveals interactions among protein networks. Science 2010;329:1038–1043.) |
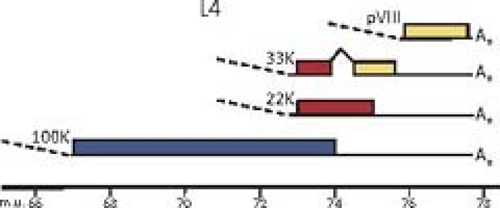
Except for the IVa2 and pIX transcription units, each of the adenovirus genes transcribed by RNA polymerase II gives rise to multiple mRNAs that are differentiated by alternative splicing, and in the case of the major late, E2, and E3 transcription units by the use of alternative poly(A) sites as well. The L4
intermediate mRNAs shown in Figure 55.8 encoding proteins L4 22K and L4 33K (the spliced mRNA) are inferred from the recent discovery of a promoter at ∼72 map units and expression of these proteins at intermediate times postinfection.226 As mentioned earlier, the analysis of adenovirus mRNA structure led to the discovery of RNA splicing24,53 (Fig. 55.9).
intermediate mRNAs shown in Figure 55.8 encoding proteins L4 22K and L4 33K (the spliced mRNA) are inferred from the recent discovery of a promoter at ∼72 map units and expression of these proteins at intermediate times postinfection.226 As mentioned earlier, the analysis of adenovirus mRNA structure led to the discovery of RNA splicing24,53 (Fig. 55.9).
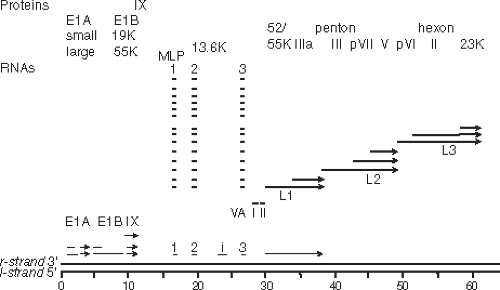 Figure 55.8. Genomes of adenoviruses type 2 and type 5. The genome of 35,937 base pairs (type 2) is divided into 100 map units. Early mRNAs are diagrammed with their exons represented by thin lines, late mRNAs with heavy lines, and intermediate RNAs (IX, IVa2, L4 intermediate, and E2A late) by lines of intermediate thickness. Arrowheads represent polyadenylated 3′ ends. Most late mRNAs originate at the major late promoter (MLP) at 16.8 map units and contain the tripartite leader whose exons are labeled 1, 2, and 3. E2B mRNAs encoding the adenovirus DNA polymerase (AdPol, 140K) and terminal protein precursor (pTP, 87K) are also expressed from the E2 late promoter and contain the same noncoding 5′ exon as the E2A late mRNAs. The L4 intermediate mRNAs encoding proteins L4 22K and L4 33K (from the spliced mRNA) are inferred from the recent discovery of a promoter at ∼72 map units and expression of these proteins at intermediate times postinfection.226 Proteins translated from the mRNAs transcribed to the right and left are named at the top and bottom, respectively. Penton represents the penton base, which is also designated virion protein III. Hexon is also designated virion protein II, and fiber, virion protein IV. pVI, pVII, and pVIII refer to precursor polypeptides that are cleaved during virion maturation. 52/55K represents a single protein that migrates as a doublet on SDS-polyacrylamide gels because of heterogeneous phosphorylation. |
Some of the protein products generated from the same transcription unit share amino acid sequence, such as the two major polypeptides encoded by the E1A region (Fig. 55.10) and the 22K and 33K proteins of the L4 region (Fig. 55.11). Others have no sequence in common, such as the two major E1B-coded proteins (Fig. 55.10) and the 100K, 22K, and pVIII proteins encoded in the L4 region (Fig. 55.11). No consistent terminology has been adopted for naming viral proteins: the early E1A proteins are named large and small E1A proteins and are often referred to by the sedimentation coefficient of the mRNAs that encode them (13S and 12S); E1B and E3 proteins are designated by their apparent molecular mass estimated from SDS gel electrophoresis; E2 proteins are named for their functions; E4 proteins are named for ORFs; and the proteins encoded by the major late transcription unit are named for the virion proteins discussed above in Virion Structure and the molecular mass for nonvirion proteins. The various historical names of viral polypeptides are used in this chapter, generally preceded by the name of the transcription unit or family of late mRNAs that encodes them (e.g., E4orf6, L4-100K).
Many of the individual adenovirus transcription units encode a series of polypeptides with related functions. As will be discussed, the E1A unit encodes two principle proteins that activate transcription and induce the host cell to enter the S phase of the cell cycle; E1B encodes two proteins that block apoptosis; E2 encodes three proteins that function directly in DNA replication; E3 encodes products that modulate the response of the host to infection; and the late family of mRNAs is concerned with the production and assembly of capsid components. Only the E4 unit encodes an apparently disparate set of functions. E4 products mediate transcriptional, RNA splicing, and translational regulation; mRNA nuclear export;
and modulate DNA replication and apoptosis. The grouping might be driven in part by the advantage of using a single transcriptional control element to regulate the expression of multiple polypeptides that are needed simultaneously to execute a function such as DNA replication. As for bacteriophage, the grouping of coding regions for products that interact physically or functionally may have been selected during virus evolution so that recombination between different viral genomes in co-infected cells did not separate genes for interacting proteins.
and modulate DNA replication and apoptosis. The grouping might be driven in part by the advantage of using a single transcriptional control element to regulate the expression of multiple polypeptides that are needed simultaneously to execute a function such as DNA replication. As for bacteriophage, the grouping of coding regions for products that interact physically or functionally may have been selected during virus evolution so that recombination between different viral genomes in co-infected cells did not separate genes for interacting proteins.
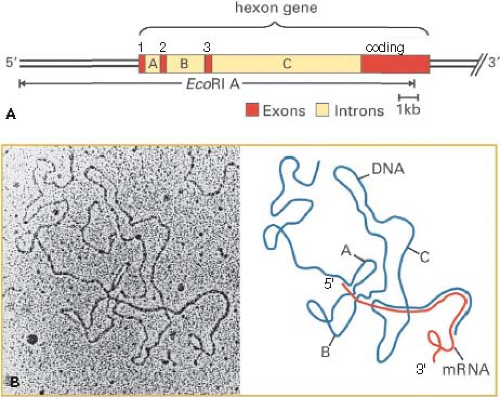 Figure 55.9. Discovery of spliced Ad2 hexon mRNA by electron microscopy of an RNA-DNA hybrid. A: Diagram of the Ad2 EcoRI A fragment and the hexon gene with exons 1, 2, and 3 comprising the 5′ tripartite untranslated leader region and the 3′ coding exon shown in red, and introns labeled A, B, and C. B: Electron micrograph of a hybrid between hexon mRNA and the EcoR1 A coding strand visualized by formamide spreading. In this method, RNA-DNA hybrid appear as a smooth filament whereas single-stranded DNA and RNA appear slightly thinner and less smooth.70 A diagram of this RNA-DNA hybrid is shown at the right with hexon mRNA in red and single-stranded DNA of the hybridized EcoRI A fragment in blue. Regions where the red RNA parallels the blue DNA represent base-paired regions of the RNA-DNA hybrid. (B, left, from Berget SM, Moore C, Sharp PA. Spliced segments at the 5′ terminus of adenovirus 2 late mRNA. Proc Natl Acad Sci U S A 1977;74:3171–3175, with permission.) |
Stages of Replication
The replication cycle is divided by convention into two phases that are separated by the onset of viral DNA replication. Early events commence as soon as the infecting virus interacts with the host cell. These include adsorption, penetration, movement of partially uncoated virus particles to a nuclear pore complex (NPC), transport of viral DNA through the NPC into the nucleus, and expression of an early set of genes. Early viral gene products mediate further viral gene expression and viral DNA replication, induce cell cycle progression (presumably to activate host cell genes for nucleotide and protein synthesis), block apoptosis, and antagonize a variety of host antiviral measures (see Chapter 56, Adenoviruses by W.S.M. Wold and M. Ison). In HeLa cells infected at a multiplicity of 10 plaque-forming units per cell, the early phase lasts ∼ 6 hours, after which viral DNA replication is first detected. Concomitant with the onset of viral DNA replication, the late phase of the cycle begins with expression of late viral genes and assembly of progeny virions. The infectious cycle is completed after 24 to 36 hours in HeLa cells. At the end of the cycle, about 105 progeny virus particles per cell have been produced, along with the synthesis of a substantial excess of virion proteins and DNA that are not assembled into virions.113 Cells infected at high multiplicity seldom divide; hence, at the completion of the replication cycle, the DNA and protein content of the infected cell has increased by a factor of about two.
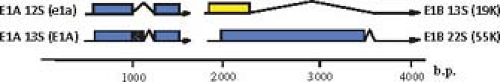 Figure 55.10. Ad2 and Ad5 major E1A and E1B mRNAs and proteins expressed during the early phase of infection and in transformed cells. The large and small E1A proteins are translated from spliced mRNAs with single introns having alternative 5′ splice sites and the same 3′ splice site (12S and 13S E1A mRNAs). Translation starts at the first AUG of both mRNAs, and the second exon of both mRNAs is translated in the same open reading frame. As a consequence, the large E1A protein (289 aa residues) contains 46 amino acids near the middle of the protein that are absent from the small E1A protein.263 Two E1B proteins of 21K (often called 19K because of its apparent molecular weight on SDS-polyacrylamide gels) and 55K are translated from the first and second AUG in alternative reading frames of the large, 22S E1B mRNA. The 21K protein is also expressed from the first AUG of the smaller 13S E1B mRNA.30 |
Studies of the human adenovirus replication cycle have focused primarily on the closely related HAdV-2 and HAdV-5 (Ad2, Ad5) viruses. These closely related types have been favored because they are easily grown in the laboratory, and
an extensive collection of mutant viruses have been developed. When other human types have been studied, their growth strategies usually have proved similar to the paradigm established for these prototypes. Most studies of adenovirus growth have been performed by infection of HeLa or KB cells at fairly high multiplicities of infection (more than 10 plaque-forming units per cell). High multiplicities of infection have been used so that all cells in the culture are synchronously infected, allowing the ordered series of biochemical events during the infectious cycle to be observed in a time-wise fashion. HeLa and KB cells have been favored as hosts because they are easily propagated in large quantities, and because Ad2 and Ad5 grow in them rapidly and to high yield. These tumor cells support more rapid viral growth than human diploid fibroblasts, in which the replication cycle is prolonged to about twice as long due to extension of the early phase of infection.107
an extensive collection of mutant viruses have been developed. When other human types have been studied, their growth strategies usually have proved similar to the paradigm established for these prototypes. Most studies of adenovirus growth have been performed by infection of HeLa or KB cells at fairly high multiplicities of infection (more than 10 plaque-forming units per cell). High multiplicities of infection have been used so that all cells in the culture are synchronously infected, allowing the ordered series of biochemical events during the infectious cycle to be observed in a time-wise fashion. HeLa and KB cells have been favored as hosts because they are easily propagated in large quantities, and because Ad2 and Ad5 grow in them rapidly and to high yield. These tumor cells support more rapid viral growth than human diploid fibroblasts, in which the replication cycle is prolonged to about twice as long due to extension of the early phase of infection.107
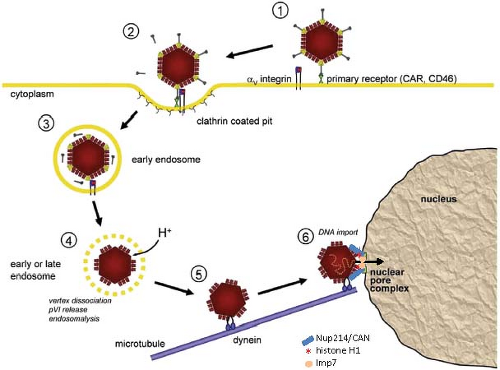
Mechanism of Attachment
Adenovirus uses distinct cellular receptors for attachment and internalization (Fig. 55.12). Initial attachment of HAdVs in species A, C, D, E, and F (but not species B) is mediated by high-affinity binding of the fiber knob domain75 to the host-cell transmembrane CAR protein (coxsackie B, adenovirus receptor).23,281,339 The CAR protein (345 residues in human) is a component of epithelial cell tight junctions with extracellular domains in the immunoglobulin (Ig) superfamily.58 It is
abundantly expressed in heart, pancreas, the central and peripheral nervous systems, prostate, testis, lung, liver, and intestine, but little or no CAR is expressed on hematopoietic cells or adult muscle.220 Infection does not require the CAR cytoplasmic or transmembrane domains,363 implying that CAR function in infection is primarily as an anchor for high-affinity binding. The finding that most CAR is localized below the apical surface of polarized epithelial cells comprising the respiratory epithelium at tight junctions on the basolateral surface raises the question of how species C respiratory HAdVs like types 2 and 5 initially infect such tissues from the lumenal side. It has been suggested that infections may initiate in specialized nonpolar cells that express CAR on their apical surface, or that lesions of the epithelium are required to expose basolateral surfaces of epithelial cells during the initial phase of infection.220
abundantly expressed in heart, pancreas, the central and peripheral nervous systems, prostate, testis, lung, liver, and intestine, but little or no CAR is expressed on hematopoietic cells or adult muscle.220 Infection does not require the CAR cytoplasmic or transmembrane domains,363 implying that CAR function in infection is primarily as an anchor for high-affinity binding. The finding that most CAR is localized below the apical surface of polarized epithelial cells comprising the respiratory epithelium at tight junctions on the basolateral surface raises the question of how species C respiratory HAdVs like types 2 and 5 initially infect such tissues from the lumenal side. It has been suggested that infections may initiate in specialized nonpolar cells that express CAR on their apical surface, or that lesions of the epithelium are required to expose basolateral surfaces of epithelial cells during the initial phase of infection.220
The x-ray crystal structure of a complex between the HAdV-12 fiber knob and the extracellular N-terminal Ig-like domain (D1) of CAR showed that three CAR molecules bind to the three interfaces between fiber knob monomers26 through the same surface used by CAR for homotypic interactions.99 The avidity generated from the three independent interactions generates high-affinity binding with a KD ≈ 1 nM.200 Free fiber proteins produced during the late phase of infection in excess over fiber proteins incorporated into virions are released with progeny virions from the basolateral surfaces of infected airway epithelial cells and interfere with CAR oligomerization at tight junctions. This probably promotes release of progeny virions to the airway lumen.359 In this way, the fiber–CAR interaction serves two functions: initial attachment of virions to host cells during infection, and dissemination of progeny virions to new host cells and host organisms.
In contrast to other HAdV species, fiber knobs of HAdVs in the B species and species D HAdV-37 bind CD46, a regulator of the complement cascade present on the plasma membrane of most cell types including hematopoietic cells.102,295,301,382 As in the case of CAR binding, three CD46 extracellular domains are bound by the three fiber monomer interfaces in the fiber knob.264
As mentioned earlier, HAd-40 and −41 incorporate one of two different fibers at each vertex of the virion. The longer of these binds to CAR. The receptor for the second, shorter fiber protein of HAdV-40 and −41 is not known, but it is postulated to contribute to the tropism of these viruses for intestinal epithelium and hence the enteritis with which they are associated.60 The stability of HAdV-40 and −41 at low pH no doubt also contributes to this tropism.89
The fiber knob of HAdV species D type 37 and probably other species D types can bind three molecules of sialic acid near its top39 as opposed to the interaction of CAR and CD46 with the lateral surface of the fiber knob. Consequently, this terminal saccharide present on many glycoproteins may also function as an initial attachment site for HAdV species D types. Infection by HAdVs elicits a potent, long-term humoral immune response. While most neutralizing antibodies bind to the major virion protein hexon and are primarily responsible for the classification of HAdVs into serotypes,104,266,278,323,340 neutralizing antibodies to fiber are also generated90 and neutralize synergistically with antibodies to penton base, which interacts with the secondary HAdV receptor.103
In addition to these well-studied host receptors, experiments with HAdV-5–based gene delivery vectors led to the discovery that most vectors infect hepatocytes following intravenous administration. This was found to result from binding of coagulation factor X through its negatively charged Gla-domain to the central depression of the Ad5 hexon and subsequent uptake into hepatocytes through an interaction with heparan sulfate proteoglycans.357 A review of other potential receptors for human adenoviruses in specific tissues is presented in.14
Mechanism of Entry, Intracellular Trafficking, and Uncoating
Following high-affinity binding of the HAdV-5 fiber knob to CAR, the RGD loop domains of penton base subunits (Fig. 55.5) associate with αvβ3 and αvβ5 integrins on the host-cell surface with 50-fold lower affinity than the interaction of fiber with CAR234,235,373 (Fig. 55.12). Integrins are abundant heterodimeric transmembrane proteins involved in cell adhesion to the extracellular matrix and neighboring cells via RGD peptide motifs in the bound, extracellular protein and are exploited by many viruses for cell entry.316 At saturation, ∼4 integrins are bound to the pentameric penton base.52 Neutralizing antibodies that bind loops on the outer surface of penton are generated that block association with integrins.137 However, a monoclonal antibody directed against the RGD loop cannot neutralize the virus, whereas Fab fragments from the same antibody can.315 This is presumably because steric hindrance due to the central fiber and the short distance (∼60 Å) between the RGD loops prevents binding of the intact antibody to all five penton base subunits.
Interaction of the penton base with integrins is presumably facilitated by their high local concentration resulting from high-affinity binding of the fiber knob to transmembrane CAR. Binding of the penton base to integrins requires the flexibility of the fiber shaft.381 Clustering of integrins through the binding of several integrins to the subunits of one pentameric penton base triggers integrin transmembrane signaling, leading to activation of phosphoinositide-3-OH kinase (PI3K), p130CAS, and Rho family GTPases.186 This results in the localized actin polymerization required for endocytosis of the virion via clathrin-coated pits in the classic receptor-mediated endocytosis that involves clathrin adapters and the large GTPase dynamin220 (Fig. 55.12). Dynamin multimerizes into a spiral-shaped structure that promotes the membrane fusion required for separating an endosome from the plasma membrane.105,201,362
In contrast to the other HAdV species, species B HAdVs, with fiber knobs that bind CD46 as the primary receptor, enter cells through a subsequent penton base interaction with αv integrins that stimulates an alternative, clathrin-independent mechanism of endocytosis called macropinocytosis.6,154 Macropinocytosis generates large vesicles near the plasma membrane and requires activation of the PAK1 protein kinase, protein kinase C (PKC), the small GTPase Rac1, Na+/H+ exchangers, and the C-terminal binding protein 1 (CtBP1). CtBP1 (C-terminal binding protein) is so named because it binds to the C-terminal conserved region 4 of HAdV E1A proteins.51 Remarkably, CtBP1 and 2 also function in the nucleus to regulate transcriptional activation by E1A (see Regulation by E1A). Species C HAdVs also stimulate macropinocytosis, but do not use this process as a primary route of infection.219 However, this phenomenon probably accounts for the observation that infection of cells with untreated or inactivated HAdV-2 or −5 can stimulate uptake of macromolecules from the medium.59,95
Uncoating of the virion begins at the cell surface. The interaction of penton base with integrins at the cell surface and during endocytosis leads to detachment of fibers, so that a fiberless virion is endocytosed.232 Structural studies revealed substantial conformational flexibility in the central region of the penton base pentamer such that an ∼15 Å diameter pore in the cryoEM structure of wild-type HAdV-5196 becomes an ∼28 Å pore in the x-ray crystal structure of the isolated penton base403 and can expand to a diameter of ∼50 Å in the crystal structure of an HAdV-5 engineered with a short fiber derived from HAdV-35 to facilitate crystallization.274 Such an expansion of the penton base central pore may facilitate release of the fiber. Studies of virion disassembly using fluorescently labeled virions and fluorescence resonance energy transfer (FRET), fluorescence anisotropy measurements of the segmental motions of fluorophores, and fluorescence lifetime imaging microscopy using a pH-sensitive probe indicated that the half-time for detachment of the fiber is 3 minutes following adsorption at 4° followed by warming to 37°.214
Following endocytosis of clathrin-coated vesicles containing HAdV-2, the vesicles mature into endosomes, and subviral particles lacking fibers escape from endosomes into the cytosol ∼15 minutes after binding to the cell surface.110 Release from endosomes may require further signaling from αv integrins following endocytosis since release is blocked by inhibitors of protein kinase C, which prevent transport of virion containing vesicles through the actin cortex under the plasma membrane into the cytosol.109,232 Release of penton base, peripentoneal hexon trimers and the internal capsid proteins IIIa, VI, and VIII occurs in the endosome, thought to be triggered in part by acidification during endosomal maturation112 (Fig. 55.12). Exposure to a reducing environment in the maturing endosome also results in re-activation of the virion-associated viral protease, which functions during virion assembly (see Virion Assembly), and cleavage of protein VI. This is required for final disassembly of subvirion particles at nuclear pore complexes and DNA import into the nucleus.111 Lysis of the endosome and release of partially uncoated virions into the cytosol is mediated by an amphipathic helical, membrane lytic domain of protein VI375 that appears to be sequestered in the intact virion in cavities on the inner surface of hexon trimers.274 Thus, the virion, which is very stable outside the cell, is dismantled by an ordered elimination of structural proteins on entry to the cell so that it can deliver its DNA to the nucleus.
For HAdV species A, B, C, and D, release of penton base and protein VI and lysis of endosomes are blocked by α-defensins, antimicrobial cysteine-rich peptides released from host neutrophils.181,239,305 CryoEM and other studies indicate that thousands of defensin molecules bind to HAdV species A, B, C, and D (but not E or F) virions. Defensins bound at the interface between fiber and penton base are particularly important for inhibiting infection by preventing the loss of the vertex region and therefore preventing escape of protein VI and endosome lysis.306 As a consequence, virion-containing endosomes mature into late endosomes and fuse with lysosomes where virion proteins and DNA are degraded.306 Anti-hexon neutralizing antibodies cross-link hexons and also may prevent the release of peripentoneal hexons as well as the subsequent release of protein VI and endosome lysis so that virions are delivered to lysosomes and degraded.
In the absence of these host defenses, endosomal lysis occurs. The subvirion particles released into the cytosol are transported to the nucleus on microtubules66,183,207 (Fig. 55.12). Fluorescently labeled virion particles in live cell imaging are observed to switch rapidly between minus-ended microtubule motility toward the juxtanuclear microtubule organizing center (MTOC) and plus-ended motility away from the nucleus. But transient activation of protein kinase A (PKA) by integrin signaling and activation of the p38 MAP kinase and its target MAPKAP kinase 2 (MK2) by an unknown mechanism increase the frequency and velocity of minus-end directed movement of subviral particles by the dynein motor protein complex324 until they reach the MTOC.18 One neutralizing monoclonal antibody to hexon does not interfere with endosome lysis, but rather blocks infection by inhibiting the association of virions with microtubules, suggesting that it is hexon that binds to microtubule motors.304
From the juxtanuclear MTOC, subvirion particles associate with nuclear pore complexes (NPCs)110 via an interaction between hexon trimers and the FG-repeat–containing domain of NPC cytoplasmic filament protein CAN/Nup214.344 Surprisingly, the activity of exportin 1 (CRM1), a nuclear export factor, is required for this association with the cytoplasmic side of NPCs since association of subvirion particles with NPCs is blocked by leptomycin B and ratjadone A, highly specific inhibitors of exportin 1, and by small interfering RNA (siRNA) to exportin 1 mRNA.321 But how exportin 1 functions in this process is unclear.
The diameter of subviral particles bound to the NPC (∼90 nm) is too large to pass through diffusional channels in the pore, ∼5 to 9 nm.4 Viral uncoating occurs at the nuclear pore as shown by exposure of core polypeptide VII and hexon epitopes that are hidden in intact virions.110 Association of intact subviral particles with NPCs occurs by ∼60 minutes postinfection, and particle disassembly assayed by epitope exposure occurs by ∼120 minutes.344 FRET experiments with fluorescently labeled virions indicate that the half-time for disassembly of the icosahedral capsid, probably coincident with unpackaging at the NPC, is 60 minutes.214 Blocking NPC binding by microinjection of wheat germ agglutinin or specific antibodies that bind to central domains of the NPC blocked exposure of the epitopes, indicating that an interaction with the NPC is required to trigger final capsid disassembly.110 This mechanism assures that viral DNA is released only at the NPC in preparation for transport into the nucleus.
Viral DNA bound by the major basic core protein VII110,310,358,386 enters the nucleus by ∼120 minutes postinfection, while most hexon and protein IX remain associated with NPCs, indicating that the viral DNA–protein VII complex separates from the partially dissociated capsid during the nuclear import process.110 Protein VII and the viral DNA to which it is bound are imported into the nucleus by a Ran-dependent mechanism involving protein VII binding by the abundant importin Transportin.135,379 In addition, free histone H1, which is known to be in rapid dynamic equilibrium with chromatin-bound H1,184,223 binds to a cluster of 16 acidic residues on one of the hexon surface loops conserved among type C HAdVs.344 Subsequent import of a small fraction of the hexon complexed to histone H1 by an Importin 7–importin β heterodimer is proposed to contribute to the nuclear import of the viral DNA-protein VII complex.344 Experiments with HAdV-2 containing a green fluorescent protein–protein V fusion in its core indicate that the core protein V remains on the cytoplasmic side of the
nuclear pore and does not enter the nucleus with the protein VII–viral DNA complex.270 Protein VII remains associated with viral DNA after its import into the nucleus until it is dissociated by transcription of the viral genome.47,358,386
nuclear pore and does not enter the nucleus with the protein VII–viral DNA complex.270 Protein VII remains associated with viral DNA after its import into the nucleus until it is dissociated by transcription of the viral genome.47,358,386
The process of HAdV infection activates several components of the innate immune response that recognize pathogen-associated molecular patterns (PAMPs). Binding of purified recombinant HAdV-5 fiber knob to the CAR receptor of human A549 cells derived from alveolar type II epithelial cells caused a sharp peak in activity of MAP kinases ERK1/2 and JNK 5 to 20 minutes after addition to cells, and NF-kB translocation to nuclei by 4 hours.327 Although the mechanism of signaling between fiber knob bound CAR and the MAP kinase and NF-kB activation pathways remains unclear, this was associated with expression of proinflammatory chemokines interleukin-8, CXCL-1 (GRO-α), CXCL-3 (GRO-γ), CCL5 (RANTES), and CXCL-10 (interferon-inducible protein 10). No activation was observed with purified penton base or hexon.327 Toll-like receptor 9 (TLR9) is activated by viral DNA released into endosomes in plasmacytoid dendritic cells, resulting in secretion of type 1 interferon α.21,42,146,399 In macrophage, following endosomal lysis by protein VI, the cytosolic NLRP3 inflammasome265 is activated by released cytosolic viral DNA. This triggers activation of caspase I, which processes pro interleukins 1β and 18, leading to their secretion.230 In mouse models of intravenous administration of defective HAdV vectors, infection of splenic macrophage led to secretion of the potent interleukin 1α.76 These and potentially other innate proinflammatory immune responses likely contributed to the well-publicized fatal systemic inflammatory syndrome in an ornithine transcarbamylase (OTC)–deficient patient following infusion into a hepatic artery of 4 × 1013 particles of an HAdV-5 gene therapy vector with a substitution of an OTC expression cassette for the viral E1A and E1B regions and deletion of E4.272
The robust humoral adaptive immune response elicited by HAdV infections in immunocompetent patients results in production of neutralizing antibodies primarily to hexon, but also to fiber and penton base as discussed previously. In addition, a cellular adaptive immune response is induced. Both cytotoxic CD8+ T-cell179,180,328,329 and memory CD4+ T-cell179,245,328 responses occur to epitopes in hexon that are highly conserved between different HAdV species. Consequently, it has been suggested that adoptive transfer of adenovirus-specific T cells from healthy adults may be useful in preventing or treating HAdV infections of multiple different types in immunocompromised patients.179
Transcription
As mentioned previously, during the infection process adenovirus DNA is transported into nuclei bound by the most abundant virion core protein, the 20-kD, highly basic protein VII.110,310,358,386 Protein VII serves the crucial role of suppressing activation of the cellular dsDNA break response (DSBR) when the viral DNA associated with protein VII first enters the nucleus.158 In the absence of such protection, the cellular MRE11-RAD50-NBS1 complex is activated by the termini of the viral DNA and degrades the ends of the genome, inhibiting viral DNA replication.158
Release of core protein VII to facilitate transcription of the infecting viral genome is stimulated by its interaction with several abundant acidic nuclear proteins, including template activating factor I.120 The normal function of this protein is to remove protamine from sperm chromatin following oocyte fertilization,216 a similar process in that small basic proteins are removed from DNA. Chromatin immunoprecipitation assays168 indicate that cellular histones replace much of protein VII early in infection by a mechanism that requires viral transcription,47 generating viral genomes bound by both protein VII and acetylated nucleosomes. The DNA of an HAdV-5–based gene therapy vector was also shown to be assembled into physiologically spaced nucleosomes containing histone H3.3, the isotype associated with transcribed cellular genes, dependent on the activity of the histone chaperone HIRA.282 Further convincing evidence for the association of host histones with adenoviral DNA early during infection came from immunofluorescence microscopy showing co-localization of phosphorylated histone H2AX with infecting viral genomes of an E4orf3-minus/E4orf6-minus double mutant that induces a dsDNA break response (see E4).158
Although early and late are convenient terms for description of events that occur during the replication cycle, the functional distinction between early and late events is often blurred. Early genes continue to be expressed at late times after infection, and the promoter controlling expression of the major late transcription unit directs a low level of transcription early after infection. Also, the viral genes encoding proteins IVa2, IX,28 L4-22K and L4-33K226 begin to be expressed at an intermediate time, thus forming a delayed-early category.
Regulation of Viral and Host-Cell Transcription by E1A Proteins
There are three main goals of early adenovirus gene expression. The first is to induce the host cell to enter the S phase of the cell cycle, providing an optimal environment for viral replication. E1A and E4 gene products play roles in this process. The second is to set up viral systems that protect the infected cell from various antiviral defenses of the host organism. The E1A, E1B, E3, and VA RNA genes contribute to these defenses, which are also discussed. The third is to synthesize viral gene products needed for viral DNA replication. All three of these goals depend on transcriptional activation of the viral genome and the induction of S-phase in noncycling cells in order to provide the precursor dNTPs required for viral DNA synthesis, NTPs and RNA processing proteins required for viral mRNA synthesis, and to activate pathways required for protein synthesis.
The E1A region is the first region to be transcribed following infection237 due to strong enhancer activity of the ∼500 bp upstream of the E1A promoter.130 Two major E1A mRNAs and encoded proteins are expressed early after infection from alternatively spliced mRNAs that use one of two 5′-splice sites separated by 138 bp and the same 3′ splice site263 (Fig. 55.10). Large E1A protein auto activates transcription of the E1A region about fivefold and stimulates transcription from the E1B, E2 early, E3, and E4 start sites more than 10- (E1B) to about 100-fold (E2early, E3, and E4).224,225,378 Activation requires a stable, highly specific interaction of the unique region of the large E1A protein, conserved region 3 (CR3), with the MED23 subunit of the human Mediator of transcription complex.35,314 Activation domain interactions with the Mediator MED23 subunit both increase the assembly of preinitiation complexes on promoters and stimulate transcription elongation.40,360 An interaction between CR3 and the major cellular nuclear acetyl transferase
p300 also contributes to early gene activation by large E1A.259a In addition, a glutamic acid-proline repeat, (EP)6, immediately C-terminal to CR3 in Ad2 and Ad5, is essential for large E1A activation of the early adenovirus promoters.320 Finally, the CR3 region of Ad5 also interacts with the lysine acetyl transferase PCAF, which may also contribute to transcriptional activation by the large E1A protein.259
p300 also contributes to early gene activation by large E1A.259a In addition, a glutamic acid-proline repeat, (EP)6, immediately C-terminal to CR3 in Ad2 and Ad5, is essential for large E1A activation of the early adenovirus promoters.320 Finally, the CR3 region of Ad5 also interacts with the lysine acetyl transferase PCAF, which may also contribute to transcriptional activation by the large E1A protein.259
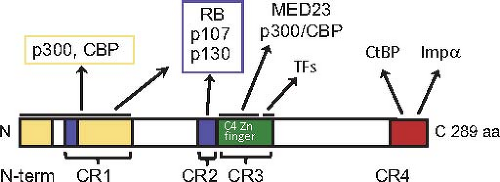 Figure 55.13. Protein–protein interaction domains of the large E1A protein. Extensively conserved amino acid sequences in the primate adenoviruses are designated CR1, 2, 3, and 4, although the N-terminal region is also extensively conserved.273 The two regions of both large and small E1A that interact with the closely related nuclear lysine amino transferases p300 and CBP are shown in yellow. The two regions that associate with the pocket domain of RB-family proteins are shown in blue. The N-terminal C4-Zn-finger region in CR3 unique to the large E1A protein that binds the MED23 subunit of the mediator of transcription complex and the C-terminal region of CR3 proposed to interact with the DNA-binding domains of host-cell transcription factors that bind the early viral promoters193 are diagrammed in green. CR4 is shown in red. It includes a C-terminal nuclear localization signal and a PXDLS sequence bound by CtBP 1 and 2 N-terminal to the nuclear localization signal.51,205 |
The small and large E1A proteins through their common sequences can each stimulate G0– and G1-arrested cells to enter S-phase in the absence of other mitogenic signals such as serum growth factors.106,139,142 Presumably, this reflects the situation when Ad2 and Ad5 initiate an infection of a human host by infecting end-differentiated, G0-arrested upper respiratory tract epithelial cells. Induction of cellular DNA synthesis requires E1A protein interactions with the abundant, closely related nuclear lysine acetyl transferases p300 and CBP (KAT3B, KAT3A)142 and the retinoblastoma family proteins RB1, RBL1 (p107), and RBL2 (p130)17,20,82,83,142,187,371,372 (Fig. 55.13). E1A proteins must form a trimeric complex between an RB-family protein, E1A, and p300 or CBP to oncogenically transform primary baby rat kidney cells.361 A model of the trimeric structure has been presented93 based on the structures of the complexes between Ad2/5 E1A residues 54 to 82 with the TAZ2 domain of CBP,93 residues 37 to 49 with the cleft between cyclin folds in the RB1 pocket domain197 and the LxCxE-containing peptide of HPV E7 protein, homologous to E1A CR2, with a complementary groove on the side of the C-terminal cyclin fold in the RB1 pocket domain178 (Fig. 55.14). The two interactions with RB-family proteins remove RB-family proteins from the E2F family of transcription factors that control genes required for entry into S-phase.17,92,197,348 The interactions of e1a with CBP and p300 and the RB-family members induce a global change in the localization of these transcriptional regulators throughout the host-cell genome.92
Stay updated, free articles. Join our Telegram channel

Full access? Get Clinical Tree