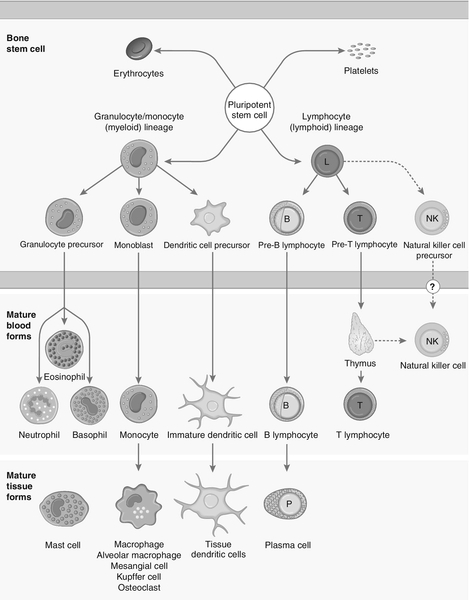
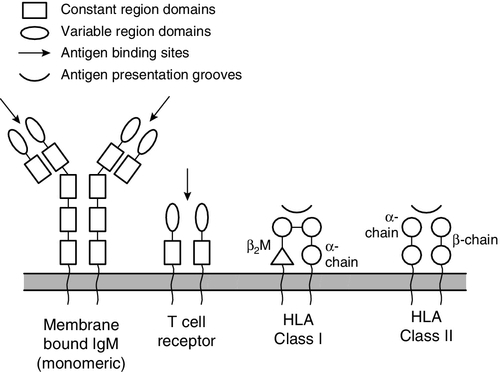
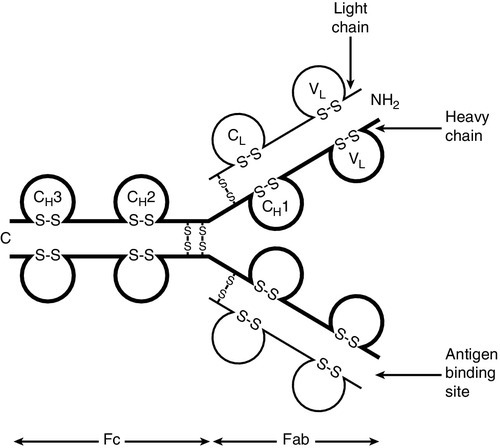
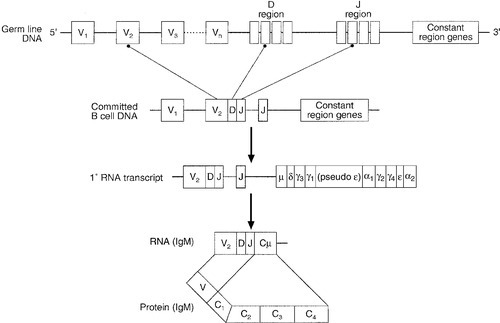
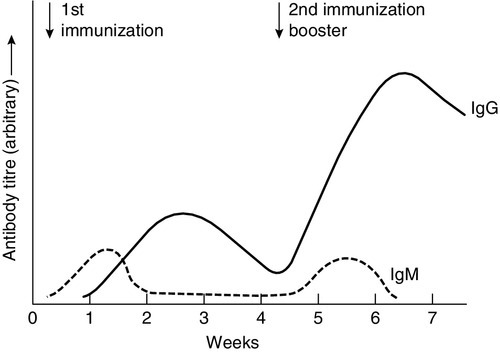
CHAPTER 30 Joanna Sheldon; Rachel D. Wheeler; Pamela G. Riches CHAPTER OUTLINE The human immune system monitors the internal and external environment at a molecular level and relays information within itself and to other systems regarding the molecular nature of the environment. Within this process, the system has evolved so that certain stimuli elicit a reaction (an immune response) while others are tolerated, eliciting no response. Microbes that invade our bodies can replicate every 20 min, giving them enormous potential to evolve more virulence. Humans reproduce approximately every 20 years with considerably less potential for population adaptation. However, an immune system that uses overlapping and interwoven processes, and can evolve by somatic mutation, allows an individual to adapt to a changing microbial environment. The immune response, like any other biological process, attempts to restore homoeostasis but may itself, if inappropriate or persistent, result in tissue damage and disease. Over the last 20 years, there has been a rapid development of our knowledge of immunology, particularly at the biochemical and molecular level, in terms of the sophisticated intercellular interactions that accompany any immune response. This chapter will make no attempt to summarize this vast body of knowledge but rather aims to convey broad concepts in describing immune responses, the immune system that makes the responses and the relevance of these to clinical laboratory medicine. The spectrum of immunological diseases includes genetic deficiencies, autoimmune diseases, allergy, malignancy, inflammation and infection. Manipulation of immune responses by inducing immune activation or suppression has great importance in many areas, including immunization and transplantation. There are two major types of immune response: innate and adaptive. The innate immune response shares many components with the adaptive response but differs fundamentally in that no specific antigen recognition is required. Instead, it involves recognition of molecular sequences that are widely expressed in nature, for example on microbial cells (lipopolysaccharide) and on damaged tissue (heat shock proteins). The adaptive response is antigen driven and requires prior antigen exposure and recognition of the antigen by T and B lymphocytes through cell surface receptors. On first contact with any antigen, the system reacts relatively slowly and transient infection may occur. Immunologically, this process is termed immunization. The initial response is known as the primary immune response: it also involves acquisition of memory of the exposure. As a result, on second contact with an antigen there is a more rapid response (the secondary immune response) and there is no infection: the host has acquired immunity. These responses involve a combination of cells, tissues and soluble mediators that are described more fully below. In order to understand immune responses, it is first necessary to consider two important terms: antigen and clonality. Most biological materials, including proteins, polysaccharides, glycoproteins and polypeptides can be antigens. How the antigen influences the immune system is related to its reaction with lymphocytes, which will determine whether it stimulates an immune response (i.e. it is an immunogen) or is tolerated (i.e. it is a tolerogen). For example, human albumin is tolerated by the human immune system but on injection into a rabbit, it will stimulate an immune response such that the rabbit produces antibodies to the protein. These antibodies, which react with human albumin, but not, for example, with horse albumin or with the rabbit’s own albumin, are described as being specific for human albumin. The antibodies produced are not directed against the entire molecule but to short, overlapping amino acid sequences termed epitopes. A large protein molecule such as albumin will have many hundreds of epitopes and, generally, the larger the protein, the more immunogenic it is. Both T and B lymphocytes have the property of antigen recognition through molecules expressed on their cell surfaces (antigen receptors). This is immunoglobulin (Ig) on B cells and the T cell receptor (TCR) on T cells. On any one cell, all the antigen receptors will be identical. Generally, this unique receptor is able to recognize one or a few epitopes. It is estimated that approximately 1010 different antigenic specificities may potentially be encountered during the lifetime of an individual, requiring approximately this number of unique receptors. The development of this very large number of lymphocyte receptors occurs before antigen contact by random rearrangements of the immunoglobulin and TCR genes. The molecular process by which this is achieved will be described in more detail in a later section. Immune responses are driven by the presence of antigen. Engagement of a compatible receptor with antigen provides signals for further proliferation and differentiation. The affinity of the antigen-receptor binding is crucial in determining the nature of that immune response. This is the molecular basis for the establishment of immune responses and immunological memory. Additionally, in the B cell, the DNA that codes for the part of the Ig receptor that binds antigen is hypermutable, so that during proliferation, somatic mutations within the receptor DNA sequences occur frequently. These mutations may result in a receptor that has a higher affinity for the antigen and, as a result, the clone bearing that receptor will compete more successfully for the antigen and receive a stronger signal, so that it is preferentially selected and responds more quickly. The secreted immunoglobulin from the B cell will similarly show this higher affinity. This process is termed affinity maturation. Immunity to many potentially lethal or harmful human diseases, for example measles, polio and smallpox, is mediated by such high-affinity immunoglobulin (antibody). One of the major roles of the immune system is to neutralize and destroy invading pathogens. These pathogens – bacteria, viruses, fungi, protozoans and worms – vary in their size, typical route of entry into the body and mechanism for evading the immune system. Generally, they enter the body across the mucosal surfaces of the respiratory system and gut and across the skin; it is in these areas of the body where the innate immune system forms the first-line of defence from invading pathogens. The major components of the innate immune system are: • mechanical barriers, e.g. intact skin, mucous membranes • sebaceous secretions; contents include fatty and other acids (hence skin acidity) • lysozyme (in tears) • gut acidity • urine acidity • intestinal motility • IGA (in tears, at mucosal surfaces) • mucus • ciliated surfaces • normal bacterial flora • acute phase proteins • interferons • other molecules (including defensins, lactoferrin, peroxidases) • neutrophils and complement, although these overlap with the acquired immune system. Intact mucosal surfaces give a high degree of protection against pathogens, and if breached, for example by surgery, burns or mechanical damage, significant infection can occur. The skin is relatively dry with a high salt content; sweat and sebaceous secretions contain fatty acids, triglycerides, lactic acid, amino acids and ammonia that have antimicrobial activity. Mucus and the cilia of the respiratory tract act together to trap and sweep inhaled particles towards the mouth and nose, where they can leave the body or where they can be swallowed to be inactivated by the gastric acid. The gastrointestinal tract has a combination of innate defence mechanisms such as the alkalinity of saliva and the strongly acidic conditions and enzymic activity of the stomach. Mucopolysaccharides in the secretions of the genital tract help make it inhospitable to pathogens. The eyes are bathed in tears containing the enzyme lysozyme, and the rate of tear production can increase to flush away any potential invading pathogens. Similarly, the urinary tract has a high flow rate that helps to keep it free from pathogens. Many areas of the body are colonized with commensal organisms. It is estimated that a normal gut is colonized with 1014 bacteria, which form the normal (commensal) bacterial flora and produce chemicals that are antimicrobial and also compete with potentially pathogenetic organisms for essential nutrients. Commensal organisms may cause infection – usually called opportunistic infections – particularly if they get into areas where they do not normally live or if the immune system is compromised. The innate immune system overlaps with the adaptive immune system (Fig. 30.1), with both systems using the complement cascade, cytokines, phagocytic cells, natural killer cells and acute phase proteins. These components can act alone in an innate response to destroy invading pathogens, but the mechanisms are non-specific and slow and are significantly enhanced by involvement of the adaptive immune system. An adaptive immune response involves an integrated system, in which there are three elements. 2. Cells with specific functions, including specific antigen recognition and presentation. 3. Soluble mediators that participate in immune reactions and can interact with other systems of the body. The components of the adaptive immune system are summarized in Table 30.1. Lymphoid tissue is distributed around the body (Fig. 30.2), with the blood and the lymphatics forming the lymphoid circulatory system. The bone marrow and the thymus are primary lymphoid organs, the bone marrow being the major site of production of all the cells of the immune system. The bone marrow is also the site of maturation of B lymphocytes. Immature T lymphocytes leave the bone marrow to develop further in the thymus (a bilobed organ in the upper anterior mediastinum that diminishes in size with age). The secondary lymphoid organs are found throughout the body, but particularly at junctions of the lymphatics, for example in the groin and axillae. This arrangement enables specialized antigen-presenting cells (APCs) to show antigens to an enormous number of lymphoid cells as they ‘traffic’ around the immune system. Secondary lymphoid tissue can be diffuse aggregates of cells or can be more organized into identifiable areas, for example the tonsils and Peyer patches in the gut. Lymph nodes also have an identifiable structure, with B cells residing mainly in the outer cortex and T cells mainly in the paracortex. Upon antigen stimulation, primary follicles within lymph nodes develop into secondary follicles with germinal centres. Activation of the immune system causes the cells within the lymph nodes to differentiate and proliferate, resulting in enlargement of the lymph nodes (lymphadenopathy). This is sought on physical examination of patients as a sign of increased activity of the immune system, for example in infection and malignancy. A large proportion of lymphoid cells are found in the gut and respiratory systems (gut and mucosal associated lymphoid tissue: GALT and MALT), where the antigenic burden is high. Although specific immune responses are made within lymphoid tissues, other non-lymphoid organs have a role to play; for example, the liver is the major site of production of the complement proteins and acute phase proteins. All cells of the body interact with the immune system. For example, any damaged nucleated cell has the capacity to make cytokines, and red blood cells have complement and antibody receptors on their surface that, owing to their enormous numbers, represent an important removal mechanism for immune complexes. Immune cells circulating in the blood are represented by the white cell population. Normally, the total white cell count of an adult is 4–11 × 109/L. This total is made up of a number of different cell types as shown in Table 30.2; the differential white cell count represents the number and proportion of each of these types. Each cell type has particular functions within the immune system: the major functions are listed in Table 30.2. TABLE 30.2 The major cells types, their approximate percentage of the differential white cell count and their major functions Each cell type has a characteristic morphology that enables it to be identified in a stained blood film examined under a microscope. Figure 30.3 shows diagrams of the various types of white blood cell with their major progenitor cell types. The cell membranes are covered with molecules that enable the cells to interact with their environment. They may be constitutively expressed (i.e. expressed in the unstimulated state) and this expression be increased on activation, or they may be induced when a cell is activated. Cells have many thousands of molecules of a whole variety of cell receptors on their surfaces. The cell surface molecules are classified by their cluster of differentiation (CD) numbers, with a different CD number for individual types of cell membrane protein. (The designation + after a CD number indicates that a specific antigen is expressed by that cell. For example, all T cells are CD3 +; T helper cells are CD4 + .) The cellular expression of these markers is used to provide a more detailed picture of cell populations, for example whether they are immature or mature, whether activated, how they are functioning and whether they are normal or malignant. Aberrant expression of the CD markers is used to classify lymphoid malignancies while reduced expression of CD antigens can be used in the investigation of immunodeficiency, for example, CD4 counts to monitor HIV infection. The technique (known as immunophenotyping), uses specific antibodies to the CD markers and flow cytometry to count and quantify cell populations. There are approximately 400 different human proteins that have been assigned a CD number. The most important immunological markers, their major functions and the cells types on which they are predominantly expressed are shown in Table 30.3. TABLE 30.3 The major CD phenotypes, their normal functions and cell types where they are normally expressed IL, interleukin; HLA, human lymphocyte antigens. The cells of the immune system arise from haematopoietic stem cells in the bone marrow. Individual stem cells respond to a number of positive or negative feedback mechanisms, for example colony stimulating factors, and are driven to proliferate and differentiate along committed pathways to give rise to populations of lymphocytes, granulocytes, APCs etc. The major processes are outlined in Figure 30.3. Neutrophils, in common with basophils and eosinophils, can be shown to contain a large number of cytoplasmic granules when stained and examined by light microscopy and are thus termed granulocytes. They have a multilobed nucleus (polymorphonuclear or PMN, hence their alternative name, polymorphs) and are the most abundant white cells in the circulation. They show high expression of adhesion molecules that enable them to roll along blood vessel walls. If expression of adhesion molecules on the vascular surface is upregulated, for example because of tissue damage or infection, neutrophils are attracted to the site of inflammation (by chemotactic factors) and leave the blood stream and enter the tissues to engulf the invading pathogens or damaged tissue components, and destroy them, in a process termed phagocytosis. The stages in phagocytosis are as follows. 2. At the site of injury, the particle or microorganism, coated with antibody and complement, adheres to the phagocyte via C3b and Fcγ receptors. 3. The adherence of the particle to the phagocyte results in the activation of the phagocyte’s membrane, initiating a change in its shape and leading to internalization of the foreign material (phagocytosis: literally, ‘eating’ the particle). 4. As the particle is taken into the cell, it becomes surrounded with a membrane, forming a phagocytic vesicle or phagosome. 5. The phagosome fuses with lysosomes to form a phagolysosome, in which the microorganism is killed, and it, or other ingested material, is digested. 6. Completely digested material is reused by the cell; incompletely digested material is released (sometimes accompanied by potentially harmful enzymes). If phagocytic cells are incubated in vitro with, for example, bacteria, phagocytosis will occur, but the process is significantly enhanced if the bacteria are coated with complement or antibody. This coating is called opsonization and, in simple terms, it enables phagocytic cells to adhere to the marked particles (giving the process specificity) and enables them to change shape to engulf the particles. Many cell types are capable of phagocytosis, the ‘professional’ phagocytic cells being neutrophils and cells of the monocyte/macrophage lineage. Many macrophages home to particular tissues where they develop highly specialized functions, e.g. Kupffer cells (in the liver), mesangial cells (kidneys), microglia (brain) and Langerhans cells (epidermis). These two cell types are also granulocytes and are particularly important in defending against multicellular pathogens, for example parasitic worms. This defence mechanism relies on the mediators in the cytoplasmic granules rather than the phagocytic functions of the cells. A close relation of the basophil, the mast cell, is tissue-based and important in allergic responses. Monocytes are immature macrophages that are travelling from their site of production in the bone marrow through the bloodstream to the tissues where they will become fixed macrophages. Potentially, they are phagocytic cells but they do not seem to act as phagocytes in the bloodstream. They have receptors that recognize many bacterial products so are very effective cells in innate immunity. Engagement of these receptors stimulates release of proinflammatory cytokines such as interleukin 1 (IL-1), IL-6 and tumour necrosis factors (TNFs). These cytokines are able to drive many aspects of inflammation and specific immunity. Monocytes therefore provide a valuable second-line defence mechanism once infectious agents have breached the mucosal defences and entered the bloodstream. The major subpopulations of lymphocytes are B and T cells. A haemopoietic stem cell first gives rise to a lymphopoietic stem cell, which in turn becomes committed to the B or T cell lineage. Lymphocytes are further divided into subsets on the basis of the presence of particular cell surface molecules (see Table 30.3). B cells (CD19 +) can differentiate into antibody (immunoglobulin)-producing plasma cells in response to exposure to antigen. T cells leave the bone marrow to mature in the thymus. They undergo further differentiation to become either T helper cells (CD4 +) that ultimately control the immune response, through cytokine production or cytotoxic T cells (CD8 +) that have an important role in killing viruses, fungi and infected cells. Other T cell types are becoming increasingly well recognized as having important immunological roles; for example, that of regulatory T cells (Treg) in the modulation of autoreactive T cells, and of Th17 (T helper) cells in inflammatory responses. There are two main types of T helper cells (Th1 and Th2), which are defined by the cytokines they produce. Th1 cells predominantly produce IL-2, TNFα/β and γ-interferon, and drive cell-mediated immunity by cytotoxic T cells, macrophage stimulation and inflammation, and production of immunoglobulins, such as IgG1 and IgG3 by B cells. Th2 cells predominantly produce IL-4, 5, 6, 10 and 13, and are important in driving IgE responses and the growth of mast cells and eosinophils. Natural killer (NK) cells (CD16 + and CD56 +) are cytolytic cells of the lymphocyte lineage, acting in a slightly different way from the cytotoxic Tc cells. B and T lymphocytes are at the heart of the adaptive immune response by virtue of their antigen receptors (Ig and T cell receptor, TCR). A comparison of the B and T cell receptors is shown in Table 30.4. The property of antigen binding is also shared by the human leukocyte antigens (HLA). These three sets of antigen-binding molecules are all products, or partial products, of the immunoglobulin gene superfamily; they are all composed of one or more homologous domains of approximately 110 amino acids, as illustrated in Figure 30.4. TABLE 30.4 Comparison of B cell receptor and T cell receptor FIGURE 30.4 The antigen presenting and binding molecules. β2M, β2-microglobulin; HLA, human leukocyte antigen. Immunoglobulins are proteins produced by cells of the B-lymphocyte lineage, either in a membrane-bound form as an antigen receptor, or as a secreted product that shows antibody activity. The membrane-bound and secreted forms differ only in the terminal amino acid residues that serve to anchor the protein into the membrane or allow secretion. The basic monomeric immunoglobulin unit consists of two identical heavy chains and two identical light chains arranged in a Y-shape (see Fig. 30.5). In humans, there are five possible heavy chains (γ, α, μ, δ and ε) that give the five immunoglobulin classes (IgG, IgA, IgM, IgD and IgE). In addition, there are four subclasses of IgG and two subclasses of IgA. There are two light chain types (κ and λ). The class (or subclass) of the heavy chains and type of light chains is determined by the common amino acid sequences in areas of the molecule called the constant domains. The light chains have one constant domain and the heavy chains have three or four constant domains, depending upon the class; it is these areas that confer functionality to secreted immunoglobulin molecules. Each chain also has a single domain of great variability in the amino acid structure, called the variable domain. The heavy and light chains are arranged such that their variable domains are adjacent, forming a groove in which antigen binding occurs. If every antigen recognition receptor on B and T cells were encoded on a one gene/one enzyme principle, these receptors alone would require more than the total DNA present within individual cells. The enormous scope for receptors to recognize antigens is achieved by a mechanism of genetic recombination. In humans, the gene for the production of the immunoglobulin heavy chain is located on chromosome 14. In the germ line, there are large numbers (estimated 250–1000) of variable (V) gene segments followed by small numbers of D (∼ 12) and J (∼ 4) gene segments. Finally, there is the constant (C) gene segment that can encode for any of the immunoglobulin heavy chains. A diagram of the immunoglobulin heavy chain gene is shown in Figure 30.6. Processing of this genetic material occurs by deletion of the introns (the non-coding DNA) and splicing together of the exons (the coding portions of the gene). One of the many V-region genes is selected and joined to one of the D-region genes, which is joined to one of the J-region genes. This VDJ sequence is then joined to the constant region genes, typically starting with the μ gene, which is the first downstream heavy chain gene in the sequence, resulting in the production of IgM. The genes for the κ and λ light chains are located on chromosomes 2 and 22, respectively, and are similar to the heavy chain gene except that they do not have a D region. Diversity of the variable region is generated by the large number of possible combinations between the V, D and J or V and J regions of the genes, variation in how these genes are spliced and by somatic mutation generated during cell development. In any cell, there is allelic exclusion, so that only one of each pair of chromosomes coding for immunoglobulin is translated. Each cell, theoretically, has two chances for each gene to be successfully rearranged. The products of unsuccessful rearrangements are secreted by the cell and degraded or cleared by the kidneys. The immunoglobulin heavy chain gene(s) is rearranged first, followed by κ-gene rearrangement. In about 40% of cells, the κ rearrangements are abortive and the cell then proceeds to λ-gene rearrangement. A small excess of free light chains (polyclonal) is produced. The immunoglobulin molecule is then assembled and expressed on the surface of B cells. The cell is now committed to a light chain type and unique variable region specificity (also called the idiotype). The organization of the heavy chain gene enables the variable region to be spliced onto a different heavy chain by the process of ‘class switching’. This occurs after interaction with antigen, when cells that show appropriate recognition of the antigen are driven to proliferate. IgM is the antibody produced in primary immune responses; IgG (or IgA or IgE), produced after class switching, is produced in secondary immune responses, as shown in Figure 30.7. Some cells will mature into memory cells and others into end-differentiated plasma cells. Plasma cells are typically non-proliferative and have little membrane-bound immunoglobulin but secrete large amounts. IgA and IgM both occur as oligomers of the basic unit. Combination is facilitated by the addition of a short polypeptide chain called the J-chain. This J-chain should not be confused with the J gene segments coding for part of the variable region of the immunoglobulin molecule – the two bear no relationship. In the plasma, IgM occurs as a pentamer. IgA can occur as a dimer and this is the predominant form in secretions. A glycoprotein called ‘secretory piece’ is synthesized by mucosal epithelial cells and integrated into secreted IgA molecules, making them more resistant to degradation by the mucosal environment. The major properties of the immunoglobulins are shown in Table 30.5. Under normal circumstances, there is enormous microheterogeneity within immunoglobulin molecules. This results in polyclonal immunoglobulins, representing all classes, light chain types and a large number of different idiotypes, each produced by a different B cell clone, even to a single antigen. Monoclonal immunoglobulins, which consist of a single heavy chain class, light chain type and idiotype, result from the proliferation of a single clone of B cells. In fact, polyclonal immunoglobulins comprise very small amounts of many thousands of monoclonal immunoglobulins. Each class and subclass of immunoglobulin has its own function or group of functions. IgG is the most abundant antibody in the circulation and also diffuses into the extravascular compartment bathing all the tissues. It is produced in secondary immune responses and is vital for immunological memory. There are four subclasses, with IgG1, IgG2, IgG3 and IgG4 being approximately 65%, 25%, 7% and 3%, respectively, of the total IgG. IgG1 and IgG3 responses mature earliest, while IgG2 and IgG4 responses mature more slowly. IgG1, IgG2 and IgG3 are all potent activators of the classical complement cascade. IgG (bound to an antigen) binds to the Fcγ receptors I, II and III on neutrophils, macrophages etc. to facilitate and enhance phagocytosis. IgG has a half-life of approximately 21 days, but this depends upon the plasma concentration, catabolism being increased with raised IgG concentrations and decreased with low IgG concentrations. IgA is the major immunoglobulin in mucosal secretions and is also produced in secondary immune responses. It is most important in binding and neutralizing organisms at mucosal surfaces, without interacting with other immunological systems. IgM is the antibody of the primary immune response, culminating in the process of class switching, inducing the B cells to make the antibodies of the secondary responses, IgG or IgA. The large pentameric structure of IgM gives good potential for antigen binding and its high molecular weight keeps it restricted to the vascular compartment. The half-lives of IgA and IgM are independent of their plasma concentrations and are approximately five days for both. IgD is detectable on B cells early in their development, but its concentration in plasma is normally very low. Its exact function in not known, but it is likely that it is important in processing antigens in immature B cells. The plasma concentration of IgE is very low; it relies on its binding to the Fcε receptor on mast cells and basophils for its activity. The release of inflammatory mediators within these cells is precipitated by binding of the surface-bound IgE to a specific antigen or allergen. The T cell receptor is somewhat simpler. It is a heterodimer in which each polypeptide chain is composed of two domains, one variable and one constant. There are four possible polypeptide chains, α, β, γ and δ. A receptor is made up of a pairing of either α with β or γ with δ. In no cell are both types expressed, nor are other pairings possible (e.g. α with δ). The variable domains are similarly constructed from gene segments, as described for immunoglobulin receptors (V, D and J for β and δ; V and J for α and γ), and then spliced into the corresponding constant regions. The T cell receptor differs from the immunoglobulin receptor in that it has no hypermutable sequences and therefore does not show affinity maturation. Also, it can only recognize antigen that is bound to an HLA (human leukocyte antigen) molecule expressed on the surface of an appropriate antigen presenting cell (APC). The major histocompatibility complex (MHC) is a group of molecules that are integral to the development of adaptive immune responses. In humans, the genes for these molecules, which are designated human leukocyte antigen (HLA), are on chromosome 6. They are expressed on the cell surface and bind peptides in an antigen-binding groove, where they are recognized by T cells via the T cell receptor. There are two types of HLA, class I and class II, summarized in Table 30.6. TABLE 30.6 Comparison of HLA class I and class II HLA molecules bind antigen but they are different from the receptors on B and T cells in that there is no genetic re-arrangement. Instead, the HLA genes are highly polymorphic, i.e. their nucleotide sequences vary between individuals giving different alleles, summarized in Figure 30.8. FIGURE 30.8 Simplified overview of the MHC locus on human chromosome 6. The MHC locus is 4 MB and encodes ∼ 300 genes. Dashed lines are used to indicate that other genes lie between the ones shown. HLA class I molecules are widely expressed on most nucleated cells. They comprise a small non-membrane polypeptide, β2-microglobulin (the invariant chain of MHC class I) covalently linked to a heavy chain, an integral membrane protein with external domains. The variation within MHC class I arises from polymorphisms within genes for the heavy chains. There are three loci for class I (HLA-A, HLA-B and HLA-C). Each individual inherits A, B and C from each parent and, unlike the situation with immunoglobulins and the TCR, there is no allelic exclusion: both sets of genes are translated. All types occur on any one cell. Because of the polymorphisms, it is likely that any individual will be heterozygous at each locus; therefore six products will be formed. HLA class II molecules are primarily expressed on immune cells, particularly those able to present antigen. They may be inducible on many other cell types. They comprise two transmembrane polypeptides (α and β chains) of similar size. As for class I, there are three loci (HLA-DR, HLA-DQ and HLA-DP). Again, the likelihood is that these properties will result in at least six distinct receptors. The class II system is, however, more complex. During evolution, there has been replication of some loci such that it is possible to express a number of different DR and DQ types, leading to increased diversity. Even with the variations described above, it is apparent that the total number of different HLA types expressed in any one individual is relatively small compared with immunoglobulins and TCRs. Unlike immunoglobulin and the TCR, HLA molecules are loaded with peptide antigen intracellularly during molecule assembly, so that prior ‘loaded’ HLA is expressed on the cell surface. Indeed, it would appear that antigen-binding is essential for stability of the molecule and that HLA without antigen is intrinsically unstable. Antigen binds in grooves formed by the folding of class I molecules and the juncture of the α and β chains in class II. The source of the antigen is different for each class. Class I molecules bind antigens synthesized intracellularly, e.g. self-peptides or viral antigens; if these antigens are able to stimulate an immune response it is via CD8 + T cells, resulting in destruction of the target cell. Class II molecules bind antigen that was derived extracellularly, e.g. microbes or microbial products. These antigens are internalized within the APC, where they are subjected to partial proteolytic degradation and then bound to the class II molecules for antigen presentation. These antigens are recognized by CD4 + T cells and stimulate an immune response. B cells can act as antigen presenting cells. Their surface immunoglobulins are able to react with free antigen molecules, forming complexes that are taken into the cells, where the antigen is degraded into peptides. These peptides can combine with HLA class II molecules to allow antigen presentation to T cells. Antigen binding alone will rarely stimulate an immune response. Some very repetitive antigen sequences, for example polysaccharides, may directly stimulate B cells. To respond to most antigens, however, both T and B cells require additional signaling from cytokines or contact with other cell types mediated through accessory or co-ligatory molecules expressed on these cells. Accessory cell interactions are many and complex and the reader is referred to dedicated immunological texts for an account of these. The complement system consists of approximately 30 plasma proteins, normally present in inactive forms (zymogens). Activation results in the generation of a cascade of enzymes that leads to activation of the terminal cytolytic pathway (membrane attack complex). Activation can occur via two major pathways, the classical pathway and the alternative pathway. In addition, complement can be activated through the lectin pathway and may be recruited by proteolytic activity generated by other physiological pathways such as coagulation. The system is inherently labile and subject to regulation by various proteins (see below). A simple diagram of the complement system is shown in Figure 30.9. The role of the complement system as a whole is to destroy invading pathogens via the membrane attack complex and opsonization but there are many other effects of complement activation, as shown in Table 30.7. During activation of the earlier components (C2, C4, C3 and C5), smaller peptides are cleaved off the zymogens. These have powerful biological activities: they are able to initiate and potentiate inflammation by increasing blood vessel permeability and attracting inflammatory and immune cells to a site of injury via a process termed chemotaxis. FIGURE 30.9 The complement system, showing the pathways for activation and their major regulatory proteins. TABLE 30.7 The major actions of components of the complement system The larger fragment formed on activation usually has the alphabetical annotation ‘b’ (e.g. C3b) and the smaller fragment, ‘a’ (e.g. C3a). Some authors use a line over a component name (e.g. C1 The alternative pathway is initiated at the level of C3, bypassing the earlier components C1, C4 and C2, which are activated in the classical pathway. C3 spontaneously degrades in tissue fluids and is rapidly inactivated. However, on activating surfaces, including damaged tissue and components of bacterial cell walls such as lipopolysaccharides, degraded C3 is stabilized and can form an enzyme to activate further C3 and to generate an enzyme to cleave C5 and thus activate the terminal sequence of the pathway. Complexes of antigens with their antibodies (immune complexes) binding C1 activate the classical pathway. C1 is a trimolecular complex of C1s, C1r and C1q. Binding of C1 liberates an enzyme C1s (the C1 esterase), which is able to cleave C4 and C2, and an enzyme formed from a complex of the larger fragments of these two molecules can activate C3. The pathway beyond this is common with the alternative pathway. Various components of bacterial cell walls are able to bind to a protein in the blood called mannose binding lectin (MBL); this complex is able to stimulate serine proteases that can directly activate component C4. Many of the complement proteins have a natural cleavage site, making them inherently unstable; therefore the presence of a number of regulatory proteins is vital to stop uncontrolled activation of the cascade. Some of the control proteins circulate while others are membrane bound; some have enzyme activity, others are simple binding proteins. The important complement regulatory proteins are shown in Table 30.8. TABLE 30.8 The major control mechanisms of the complement cascade Inflammation is a fundamental pathological process that occurs in response to tissue injury. The inflammatory response has three main stages: local reactions; destruction and/or removal of the injurious material, and repair and healing of the damaged tissue. Stimulators of inflammation include trauma, infection, infarction and deposition of immune complexes; these result in the release of inflammatory mediators (see Table 30.9) and the classic signs of inflammation – redness, swelling, warmth or heat, pain and loss (or impairment) of function. These features may be localized, but in more severe inflammation there is a widespread response and the features also include fever, leukocytosis and the production of a variety of liver-derived acute phase proteins. The functions of these proteins are summarized in Table 30.10. TABLE 30.9 Inflammatory mediators and their actions TABLE 30.10 The functions of acute phase proteins The acute phase proteins show diverse properties, particularly in the kinetics of their responses, as shown in Table 30.11. The magnitude of response is generally related to the degree of injury, although other factors such as catabolic rate, hormonal influences and genetic variability are also important. These are biological signalling peptides, whose many actions include the control of the differentiation of white blood cells and the modulation of the actions of the cells of the immune system. They include the interleukins, interferons, tumour necrosis factors and various growth factors. They are summarized in Table 30.12. TABLE 30.12 The major groups of cytokines a RANTES: regulated on activation, normal T expressed and secreted; Th, T helper. In general, cytokines, like hormones, act through high-affinity cell surface receptors that can be widely distributed on many cell types or can be restricted to one or two cell types. The receptors can be up- or downregulated or induced, depending upon the prevailing conditions. Most cytokines are glycated and their binding to the receptors may be influenced by the configuration of the sugar residues and by the other cytokines being released in the vicinity. Most cytokines are multifunctional molecules with diverse biological actions upon a wide variety of target cells. Cytokines, in general, show significant overlap in their functions, with any one function typically being shown by a number of cytokines (pleiotropism). They mostly act locally in either an autocrine or paracrine fashion; very few, for example IL-6, show a true endocrine function. The major differences between cytokines and hormones are shown in Table 30.13. Genetic deficiencies of cytokines are rare, possibly reflecting their vital role in the maintenance of health. TABLE 30.13 Major differences between cytokines and endocrine hormones Many cytokines interact with each other and the effect of a cytokine may change depending upon the prevailing micro-environment. It is important to stress that cytokine synthesis, including that of the proinflammatory cytokines, is a normal response to injury. Their functions should not be thought of as exclusively harmful, resulting in tissue damage or systemic shock. In the majority of instances, cytokines act to coordinate the elimination of invading organisms or removal of damaged tissue, thus avoiding excessive stimulation of specific immunity that could lead to hypersensitivity reactions. The cytokines interleukin (IL)-1β, IL-6 and tumour necrosis factor alpha (TNFα) are all central to the initiation of the inflammatory response. However, they do not function in isolation: other cytokines implicated in the inflammatory response include IL-8, 10, 11, 12, 17, 18, 23 and interferon γ. In addition, both defensive and injurious effects will involve the release of many mediators of inflammation such as peptides, for example the complement fragment C5a, and lipids, for example platelet activating factor. Many of these mediators act synergistically and also induce each other’s production and the production of other cytokines, resulting in both positive and negative feedback control pathways. Cytokines are also responsible, either directly or indirectly, for healing and successful resolution of the inflammatory insult. Severe inflammatory responses frequently overlap with specific immune responses, as they are often induced by infection and, furthermore, the tissue damage accompanying severe trauma may lead to secondary infection. Immune responses are usually well controlled and host damage is minimal and reversible. Under some circumstances, resolution does not occur and an exaggerated or persistent response leads to irreversible host tissue damage and, in extreme situations, even death. These inappropriate reactions are termed hypersensitivity reactions. These are classified into four types (I-IV), each with a different mechanism. Type I hypersensitivity reactions, central to allergic responses, may occur without involvement of any other type of hypersensitivity reaction. Most reactions, however, involve more than one type of response and do not, alone, define a pathogenetic mechanism that underlies a group of diseases. There are also other reactions that do not fit neatly into this classification, for example stimulatory reactions, such as when specific immune cells are stimulated by external agents, and when autoantibodies have a stimulatory effect, for example as occurs with thyroid receptor antibodies in Graves disease. Hypersensitivity reactions result in the release of inflammatory mediators. Some mediators have direct effects on local, or even distant, tissues; others recruit and activate effector cells that further contribute to tissue damage. Type I hypersensitivity reactions are IgE mediated. The IgE antibodies are formed to an antigen (or allergen), with an individual’s tendency towards making IgE being determined by many factors including genetic, T cell responsiveness and antigenic burden. The IgE binds to high-affinity IgE receptors on the surfaces of mast cells and basophils, and these cells are now primed to react the next time the cells come into close proximity with the allergen. The cross-linking of IgE on the cell surfaces causes rapid cellular degranulation and liberation of a number of chemical mediators. The mediators released by mast cell degranulation include the preformed molecules histamine, protease enzymes, proteoglycans (heparin) and chemotactic factors. Reaction of antigen with IgE on mast cells also stimulates synthesis and release of platelet activating factor (PAF), leukotrienes (B4, C4 and D4) and prostaglandins (mainly PGD2). The mediators of type I hypersensitivity reactions are shown in Table 30.14. TABLE 30.14 Mediators of type I hypersensitivity reactions The actions of histamine depend on the site of release. In the airways, it induces smooth muscle contraction; in the skin, it causes the hallmark wheal and flare response. Widespread activation of mast cells leads to systemic effects of circulatory shock, hypotension, collapse, chest tightness and, in the most severe cases, respiratory arrest and death: this is anaphylactic shock. Type I hypersensitivity reactions occur rapidly (within approximately 20 min of an insult) and are also called ‘immediate hypersensitivity reactions’. The important characteristic of type II hypersensitivity reactions is that the antigens involved are localized to the membranes of the target cells; they may be synthesized cell products, cell membrane components or bound foreign molecules (e.g. a drug). The circulating antibodies (IgG or IgM) bind to the cell-bound antigen and activate complement. Full activation of the complement pathway results in the membrane attack complex (C5–9) being assembled on the cell surface, resulting in target cell lysis. Partial activation via C3 will also render the cell a target for phagocytosis, via C3b and IgG that bind to appropriate receptors on phagocytic cells. Activation of phagocytic cells leads to release of further tissue-damaging enzymes. The tissue damage persists for as long as antibody and antigen are present. Some of the more important type II responses involve red blood cell antigens (including transfusion reactions and haemolytic disease of the newborn) and autoantibodies to cell surface components, for example autoimmune haemolytic anaemia (in which the target is red blood cells), idiopathic thrombocytopenia purpura (in which the target is platelets) and Goodpasture syndrome (in which the target is the kidney glomerular basement membrane).
Immunology for clinical biochemists
THE IMMUNE SYSTEM
Introduction
Immune responses
Antigens
Clonality
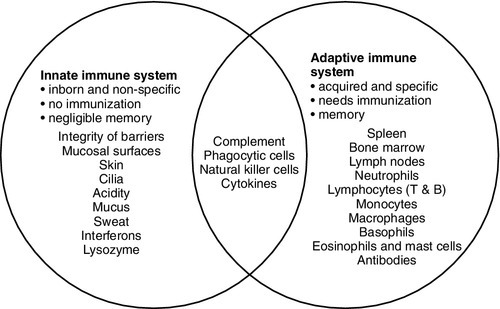
The innate immune system
The adaptive immune system
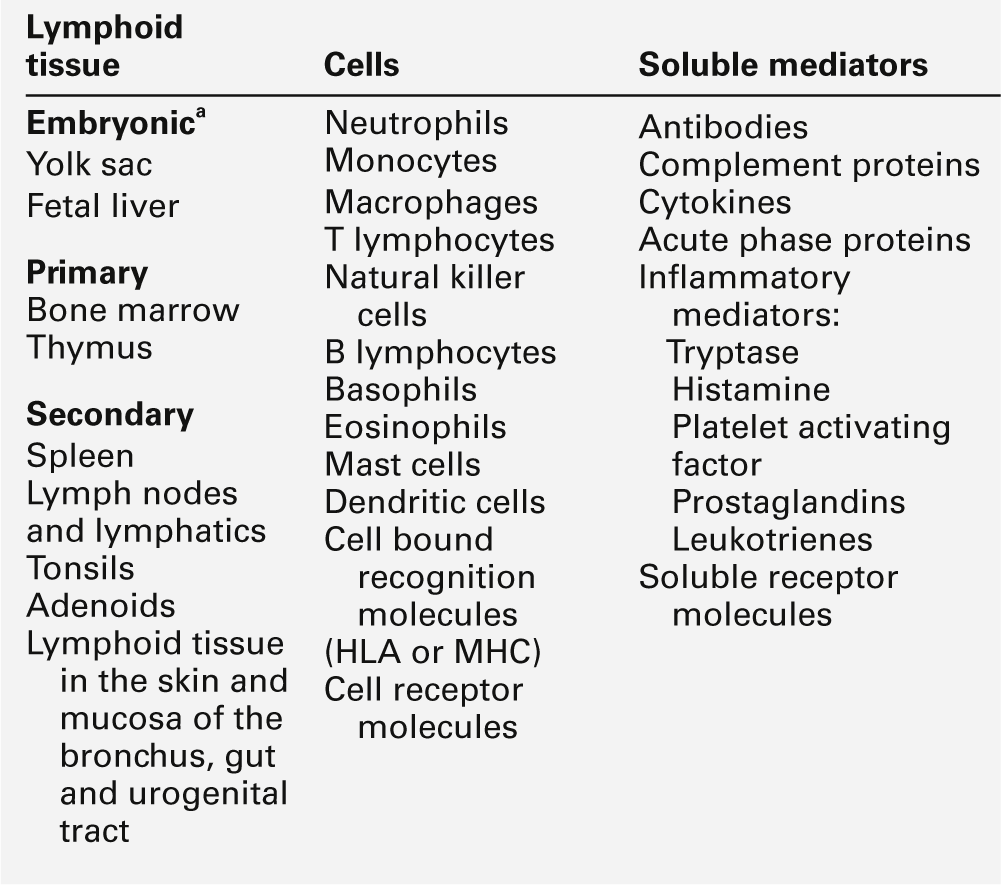
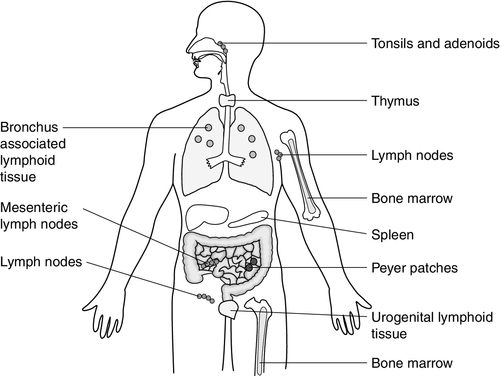
Lymphoid tissue
Cells
Cell type
Reference range (× 109/L)
Function
Total white cell count
4–11
Neutrophil
1.6–7.5
Attracted to site of inflammation by chemotactic factors
Most important cell type for phagocytosis and killing of bacteria
Opsonization with antibody and complement enhances phagocytosis
Lymphocyte
1.5–4.0
Key role in orchestrating the immune response and producing soluble mediators of immunity
T cell (CD3 +)
0.8–2.66
T helper cells (CD4 +) control the development of an immune response by secreting cytokines
Cytotoxic T cells (CD8 +) can directly kill cells, e.g. that are infected with virus
Regulatory T cells modify immune responses
B cell (CD19 +)
0.1–0.6
Antibody production
Natural killer cell (CD56 +, CD16 +)
0.05–0.60
Can directly kill cells, e.g. infected with virus, tumour cells
Monocyte
0.1–0.8
Important phagocytic cells and can act as antigen presenting cells
Cytokine production – particularly in inflammation
Basophil
< 0.1
Major role is in allergic responses via release of mediators, e.g. histamine
May be important in defending against parasites
Eosinophil
0.04–0.45
Some phagocytic function
Major role in hypersensitivity reactions via release of variety of mediators
Activation of mast cells
Phenotype
Function of protein
Cells where expressed
CD3
Part of the T cell receptor
All T cells (pan T cell marker)
CD4
Helps T cells to hold on to antigen-presenting cells (HLA class II)
T helper cells
CD8
Helps T cells to hold on to antigen-presenting cells (HLA class I)
Cytotoxic T cells
CD19
B cell co-receptor
B cells
CD20
B cell activation and differentiation
B cells
CD45
Cell differentiation, signal transduction and lymphocyte activation
All white blood cells (leukocyte common antigen)
CD16
Binds Fc γ RIII (binds IgG Fc)
Natural killer cells, neutrophils, monocytes
CD32
Binds Fc γ RII (binds IgG Fc)
Neutrophils, macrophages, B lymphocytes, eosinophils
CD64
Binds Fc γ RI (binds IgG Fc)
Monocytes, macrophages, activated neutrophils
CD89
Binds Fc α R (binds IgA Fc)
Monocytes, macrophages, neutrophils
CD21
CR2 (binds C3d)
B cells (some)
CD35
CR1 (binds C3b, C4b)
Neutrophils, monocytes, B cells, erythrocytes
CD25
IL-2R α chain
Activated T and B cells
CD56
Natural killer cells
CD40
Binds to CD40 ligand (CD154); B cell proliferation and class switching
B cells, antigen-presenting cells
CD11a (+ CD418)
Cell adhesion (to other cells)
Leukocytes
CD11b (+ CD18)
Cell adhesion (to other cells) CR3
Monocytes, neutrophils, natural killer cells
CD11c (+ CD18)
Cell adhesion (to other cells) CR4
Monocytes, neutrophils, natural killer cells, some T and B cells
CD18 (+ CD 11 a, b and c)
Cell adhesion (to other cells)
Leukocytes, monocytes, neutrophils, natural killer cells, some T and B cells
CD62E
E selectin cell adhesion (to other cells)
Endothelium, platelets
CD62L
L selectin cell adhesion (to other cells)
Neutrophils
CD62P
P selectin cell adhesion (to other cells)
Endothelium
B cell proliferation and class switching
Activated T cells
Neutrophils
Basophils and eosinophils
Monocytes
Lymphocytes
B cell receptor (membrane-bound IgM)
T cell receptor
Similarities
Variation created by gene rearrangement
Part of a protein complex including accessory chains and signalling adaptors
Variable and constant domains
Differences
Two heavy chains and two light chains
Two polypeptide chains (αβ or γδ)
Somatic hypermutation allows affinity maturation
No somatic hypermutation
Class switching from IgM to other immunoglobulin classes
No class switching
Binds directly to specific antigen
Binds to specific antigen only when bound by appropriate HLA molecule
Antigen recognition
Immunoglobulins
T cell receptors
Human leukocyte antigens (HLA)
Class I
Class II
Structure
Variable heavy chain + invariant chain (β2-microglobulin)
Two variable chains, α and β
Expression
Most nucleated cells
Antigen presenting cells; may be induced on other cell types
Antigen presented
Intracellular antigen, i.e. viral, self
8–9 amino acids
Extracellular antigen, e.g. microbes or their products
14–22 amino acids
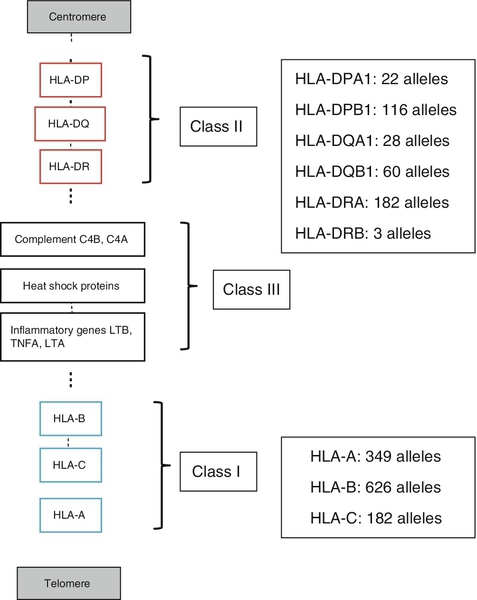
Gene loci
HLA-A, B, C
HLA-DQ, DR, DP
Responding T cell
CD8 + T cells
CD4 + T cells
Antigen presentation
Cellular immune activation
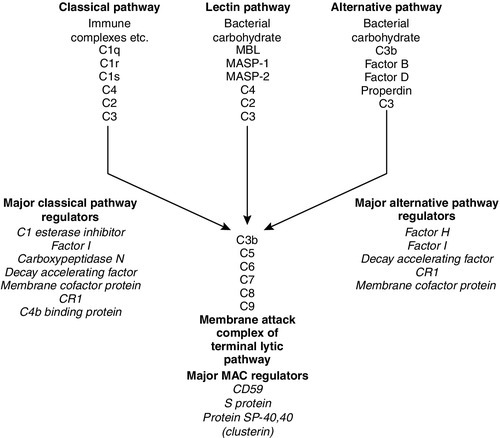
Complement
The complement system
Complement components
Effect
C1q, C3, C4
Clearance of immune complexes
Clearance of cell debris
C3a, C4a, C5a (anaphylatoxins)
Histamine release from mast cells
Increase smooth muscle contraction
Increase vascular permeability
Inflammation
C3b (opsonin)
Binding of antigen–antibody complex to receptors on phagocytes, enabling effective ingestion and destruction
C5a, C3a (chemotaxins)
Recruitment of phagocytes into areas of inflammation
C5–C9 (membrane attack complex)
Cell or bacterial lysis
Cell signalling
![]() ) to denote an active enzyme that can activate the next component; this convention is unnecessary and has not been adopted in this chapter.
) to denote an active enzyme that can activate the next component; this convention is unnecessary and has not been adopted in this chapter.
Activation via the alternative pathway
Activation via the classical pathway
Activation via the lectin pathway
Regulation of the complement pathways
Complement control mechanism
Activity
C1 esterase inhibitor
Protease inhibitor that blocks the activity of C1 esterase
Factor H
Binds C3b and enhances the degradative action of Factor I
Factor I
Enzyme that degrades C3b and C4b
C4b binding protein
Binds to C4 and enhances its destruction by Factor I
Protein S and SP-40,40 (clusterin)
Binds C5b67 and prevents formation of the membrane attack complex
Carboxypeptidase N
Enzyme that inactivates C3a, C4a and C5a
Decay accelerating factor
Transmembrane glycoprotein on most blood cells that binds C4b and inhibits C3 convertase
Membrane cofactor protein (CD46)
Membrane-bound C3 binding protein
Membrane attack complex inhibitory factor (CD59)
Membrane-bound protein that hinders insertion of the membrane attack complex
Complement receptor I
(CRI, CD35)
High affinity receptor for C3b and C4b on surface of erythrocytes
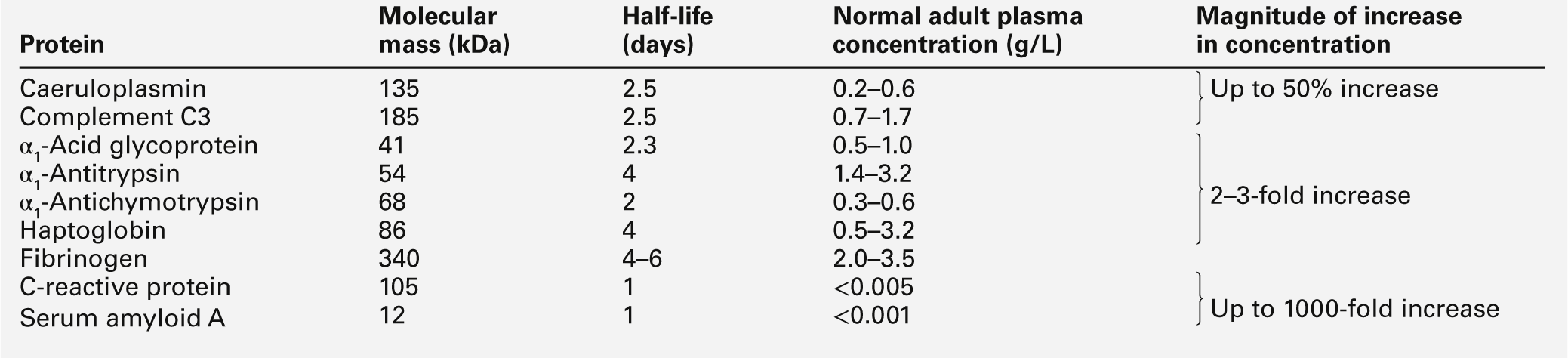
Acute phase proteins
Mediator
Action
Platelet activating factor, histamine, prostaglandins
Vasodilatation
Platelet activating factor, histamine, complement C3a and C5a, bradykinin, leukotrienes
Increased vascular permeability
IL-8, IL-1, and TNFα, complement C5a, leukotrienes
Leukocyte adhesion and chemotaxis
Bradykinin and prostaglandins
Pain
Enzymes (proteases) and products of neutrophil respiratory burst (free radicals)
Tissue damage
Inflammatory cytokines IL-1, IL-6 and TNFα
Hepatic production of acute phase proteins
Acute phase function
Examples
Mediating
Working as parts of networks of inflammatory mediators:
C-reactive protein (CRP) binds a variety of ligands and activates complement;
complement components are important opsonins and chemotactic factors;
fibrinogen and the clotting components form clots and fibrin matrices as a basis for repair
Inhibiting
Inhibiting protease activity and controlling pathways:
α1-antitrypsin and α1-antichymotrypsin inhibit the actions of enzymes released from leukocytes during phagocytosis;
C1 esterase inhibitor inhibits part of the complement system
Scavenging
Inhibiting or eliminating noxious substances produced during the inflammatory process:
haptoglobin combines with free haemoglobin to form a complex that is rapidly cleared by the liver;
CRP may opsonize DNA and cell membrane debris
Regulating
Modulating the immune response:
α1-acid glycoprotein is expressed on lymphocyte cell membranes
Repairing
Control and laying down of connective tissue elements:
α1-antitrypsin and α1-antichymotrypsin are deposited in a sequential fashion on the surface of newly formed elastic fibres;
α1-acid glycoprotein promotes fibroblast growth
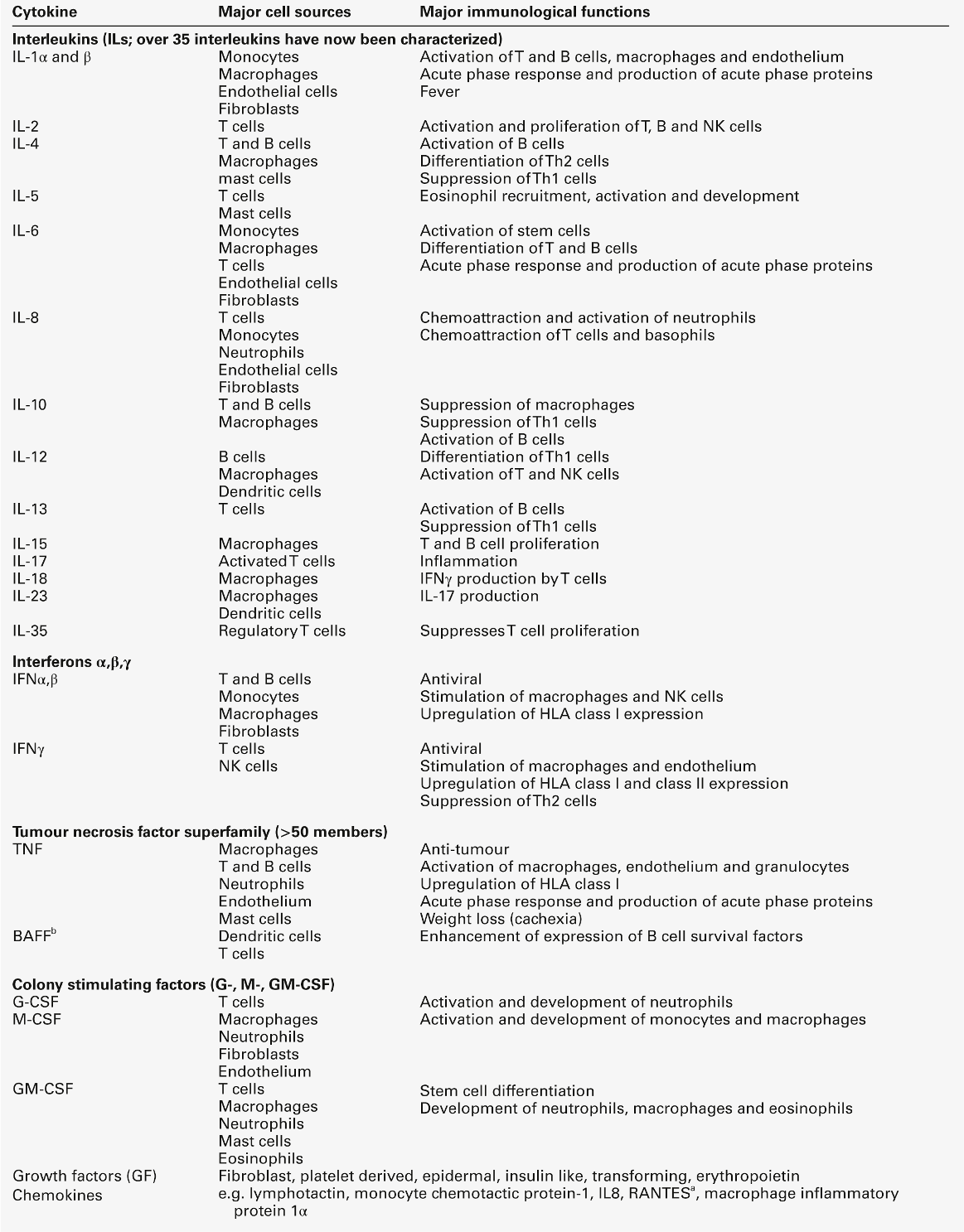
Cytokines
Property
Cytokine
Hormone
Sites of production
Many and varied
Many, but specific to each hormone
Cellular targets
Few
Many
Biological role
Fighting infection, tissue repair
Homoeostasis
Biological redundancy
High
Low
Biological pleiotropy
High
Low
Present in the circulation
Rarely
Yes
Sphere of influence
Predominantly autocrine/paracrine
Widespread and distant from site of production
Inducers
External insults
Primarily physiological changes (except stress hormones)
Inflammatory cytokines
Mechanisms of immunological damage
Type I hypersensitivity
Mediator
Pharmacological effect
Histamine
Vasodilatation, capillary permeability, bronchoconstriction
Heparin
Control of histamine release
Leukotrienes (various)
Bronchoconstriction, airway tissue oedema
Prostaglandins (various)
Potent mediators of inflammatory response
Platelet activating factor
Platelet aggregation
Tryptase
Proteolytic enzyme activates C3
Kininogenase
Kinins → vasodilatation → oedema
Cytokines (IL-5, IL-8, TNFs)
Chemoattractants
Type II hypersensitivity
Type III hypersensitivity
Stay updated, free articles. Join our Telegram channel

Full access? Get Clinical Tree