6.1 INTRODUCTION TO TRANSCRIPTION, TRANSLATION, AND TRANSPORT
In this chapter we deal with three terms derived from the Latin trans, meaning across. For our purposes, transcription refers to the writing across of genetic information from a sequence of bases in a nucleic acid to the complementary sequence in messenger RNA (mRNA), while translation converts the genetic information from the language of bases in nucleic acids to the language of amino acids in proteins. Transcription and translation are steps 3 and 4 of our generalized replication cycle (Section 5.1).
We also discuss in this chapter the transport of virus proteins and RNAs to particular locations in infected cells. We start with an overview of virus transcription, and then we discuss these three “trans” processes in eukaryotic cells. At the end of the chapter we point out some aspects of the processes that are different in bacterial cells.
6.2 TRANSCRIPTION OF VIRUS GENOMES
We have seen how there are four main categories of virus genome: dsDNA, ssDNA, dsRNA and ssRNA (Section 3.2). Because of distinct modes of transcription within the dsDNA and ssRNA categories a total of seven classes of viruses can be recognized (Figure 6.1).
Figure 6.1 Transcription of virus genomes. (+) RNA and (+) DNA have the same sequence as the mRNA, and (–) RNA and (–) DNA have the sequence complementary to the mRNA (except that in DNA thymine replaces uracil). (+) and (–) strands are not indicated for the dsDNA of the Class I viruses as the genomes of most of these viruses have open reading frames (ORFs) in both directions. (+) and (–) strands are indicated for the ssDNA of the Class II viruses. Most of these viruses have either a (+) or a (–) strand genome. A (+) RNA genome (dark blue) has the same sequence as the corresponding mRNA (green); the molecules are shown in different colors to indicate their different functions. In Class VII viruses the (+) RNA shown in blue (pregenome RNA) functions as a template for synthesis of DNA, some of which is used as a template for further transcription. Some ssDNA viruses and some ssRNA viruses have ambisense genomes. This means that that the polarity of the genome is part (+) and part (–).
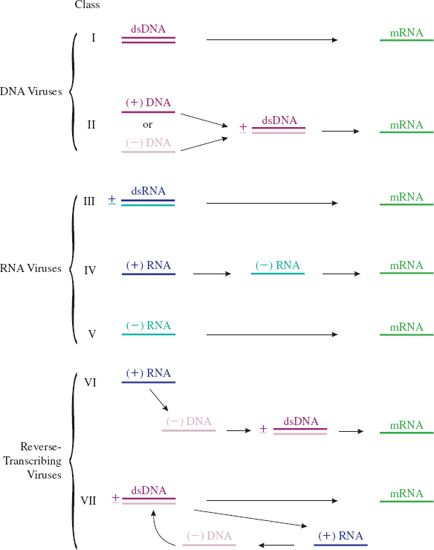
This division of the viruses into classes based on genome type and mode of transcription was first suggested by David Baltimore and this scheme of virus classification is named after him. He initially proposed six classes.
In the summary of the scheme depicted in Figure 6.1 most of the nucleic acid strands are labeled (+) or (–). This labeling is relative to the virus mRNA, which is always designated (+). A nucleic acid strand that has the same sequence as mRNA is labeled (+) and a nucleic acid strand that has the sequence complementary to the mRNA is labeled (–).
The viruses with (+) RNA genomes (Classes IV and VI) have the same sequence as the virus mRNA. When these viruses infect cells, however, only the Class IV genomes can function as mRNA. These viruses are commonly referred to as plus-strand (or positive-strand) RNA viruses. The Class V viruses are commonly referred to as minus-strand (or negative-strand) RNA viruses.
Class VI viruses must first reverse-transcribe their ssRNA genomes to dsDNA before mRNA can be synthesized. Because they carry out transcription in reverse (RNA to DNA) Class VI viruses are known as retroviruses. The ability of some DNA viruses to carry out reverse transcription was discovered later; these viruses became known as pararetroviruses and Class VII was formed to accommodate them.
There are a few single-stranded nucleic acids of viruses where there is a mixture of (+) and (–) polarity within the strand. Genomes of this type are known as ambisense, a word derived from the Latin ambi, meaning “on both sides” (as in ambidextrous). Examples of ambisense genomes include the ssDNA genomes of the geminiviruses, which are plant viruses, and the ssRNA genomes of the arenaviruses, which are animal viruses and include the causative agent of Lassa fever.
6.2.1 Modifications to the central dogma
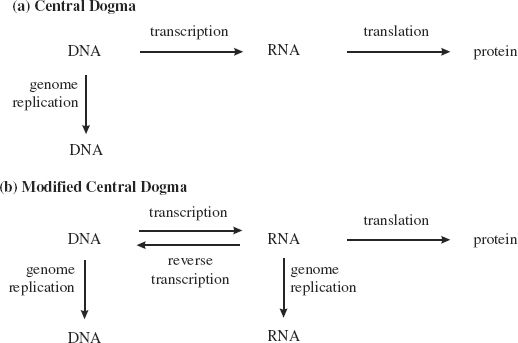
In 1958 a “central dogma of molecular biology” was proposed by Francis Crick, along with James Watson, one of his collaborators in deducing the structure of DNA. The dogma stated that the flow of genetic information is always from DNA to RNA and then to protein, with genetic information transmitted from one generation to the next through copying from DNA to DNA (Figure 6.2(a)). Increasing understanding of how viruses replicate their genomes necessitated some modifications to this dogma in 1970 (Figure 6.2(b)). Many viruses have RNA genomes that are copied to RNA, some viruses copy from RNA to DNA, and it is now known that cells can copy from RNA to DNA.
Figure 6.2 (a) Francis Crick’s central dogma of molecular biology, which proposed that genetic information is transmitted from DNA to RNA to protein, and from DNA to DNA. (b) Modifications to the central dogma, required mainly because of the various modes of virus transcription and genome replication.
6.3 TRANSCRIPTION IN EUKARYOTES
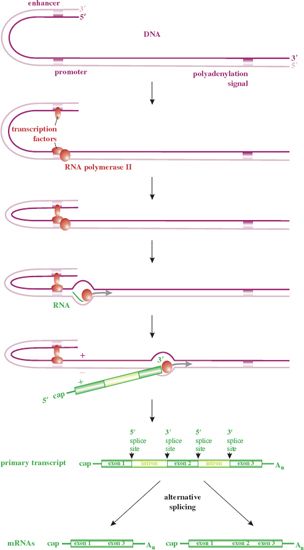
We start this section with a brief summary of transcription from eukaryotic cell genes, as many viruses transcribe their genes by similar processes, some of them using parts of the cell transcription machinery (Figure 6.3). The expression of a gene is controlled by sequences in the DNA, including:
- a promoter – binds transcription factors and the RNA polymerase;
- enhancers – bind transcription factors that significantly boost the rate of transcription.
Figure 6.3 Transcription from dsDNA in eukaryotes. Transcription is initiated after transcription factors bind to sequences within the promoter and enhancer(s). The primary transcript is capped at the 5′ end and polyadenylated at the 3′ end. The mRNAs are formed by removal of introns from the primary transcript.
6.3.1 Promoters and enhancers
The promoters of many eukaryotic cell and virus genes contain the consensus sequence:
T A T A A/T A A/T A/G
A/T and A/G indicate alternative nucleotides at those sites.
The sequence is known as a TATA box and is usually located 25–30 bp upstream from the transcription start site. A TATA box is present in the single promoter of HIV-1 (Figure 6.4), but in only one of the four promoters of hepatitis B virus (Section 19.8.3).
Figure 6.4 Part of the long terminal repeat in the HIV-1 genome, indicating the locations of some of the sequences that control transcription, and the transcription factors that bind to those sequences. The promoter (TATA box) binds TFIID, and enhancers bind:
• AP1 (activator protein 1)
• NF-κB (nuclear factor κB)
• Sp1 (stimulatory protein 1).
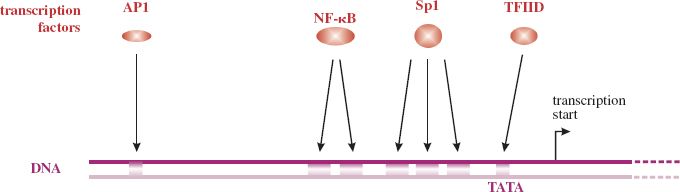
Enhancers contain sequences that bind transcription factors that may then interact with transcription factors bound to the promoter. These interactions may increase the rate of transcription starts by RNA polymerase II. Many enhancers are found only in specific cell types, for example liver cells.
6.3.2 Transcription factors
Transcription factors are proteins that bind to specific promoter and enhancer sequences to control the expression of genes. Many transcription factors activate genes, but some play other roles, including repression of gene expression.
Transcription factor IID (TFIID) is an example of a cell transcription factor. TFIID is a complex of 13 proteins, one of which is the TATA box binding protein. After TFIID has bound to the TATA box other transcription factors (TFIIA, IIB, IIE, IIF, and IIH) and RNA polymerase II bind.
Many viruses use cell transcription factors to activate or repress transcription of their genes. An example is shown in Figure 6.4, which depicts part of the HIV-1 genome in its DNA form (Chapter 18). The promoter includes a TATA box and there is a large number of enhancers.
Tissue-specific transcription factors are required by certain viruses that are specific to a particular tissue, such as liver. Some viruses produce their own transcription factors, such as herpes simplex virus VP16, which is a component of the virion (Section 11.5.2), and human T-lymphotropic virus 1 Tax protein (Section 23.10.5).
All organisms regulate expression of their genes. A frog has different genes switched on depending on whether it is in the embryo, tadpole or adult stage. Similarly, a virus may have different genes active at different stages of its replication cycle. For some viruses there are two phases of gene expression (early and late), while for others three phases (e.g. herpesviruses; see Section 11.5.2) or four phases (e.g. baculoviruses) can be distinguished.
6.3.3 Transcriptases
Transcriptase is a general term for an enzyme that carries out transcription. Most viruses encode their own transcriptase (Figure 6.5).
Figure 6.5 Enzymes used by viruses of eukaryotes to transcribe their genomes to mRNA. A (+) RNA genome has the same sequence as the virus mRNA. A (–) RNA genome has the sequence complementary to that of the virus mRNA. Transcription from DNA in the nucleus applies not only to those DNA viruses that replicate in the nucleus, but also to the reverse transcribing viruses. Some minus-strand RNA viruses transcribe in the cytoplasm and some in the nucleus.
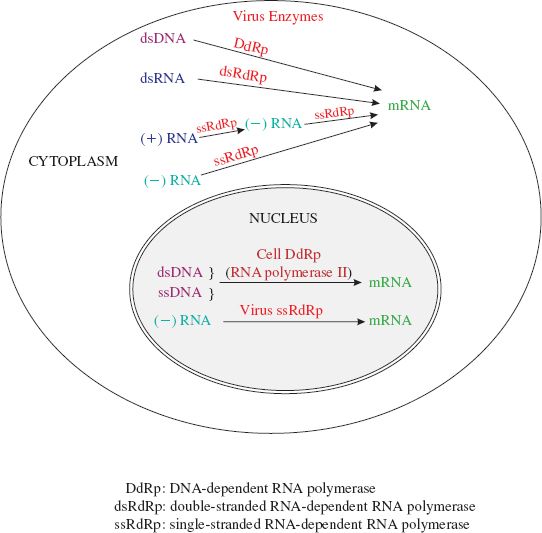
A DNA virus needs a DNA-dependent RNA polymerase to transcribe its genes into mRNA. Viruses that carry out transcription in the nucleus generally use the cell RNA polymerase II; these include the retroviruses and many DNA viruses. DNA viruses that replicate in the cytoplasm use a virus-encoded enzyme because there is no appropriate cell enzyme in the cytoplasm.
An RNA virus (apart from the retroviruses) needs an RNA-dependent RNA polymerase to transcribe its genes into mRNA. Each virus in classes III, IV, and V encodes its own enzyme, in spite of the fact that the cells of many eukaryotes encode ssRNA-dependent RNA polymerases.
Most viruses that encode their own transcriptase have the enzyme in the virion so that it is available to transcribe the virus genome when a cell is infected. Exceptions are the plus-strand RNA viruses, which must first synthesize the enzyme before transcription can begin.
The retroviruses and the pararetroviruses perform reverse transcription (Section 6.2) using enzymes known as reverse transcriptases. These enzymes are RNA-dependent DNA polymerases, but they also have DNA-dependent DNA polymerase activity, as the process of reverse transcription involves synthesis of DNA using both RNA and DNA as the template.
6.3.4 Capping transcripts
Soon after RNA synthesis has begun, and while transcription is continuing, most transcripts are “capped” at the 5′ end (Figure 6.3). The cap is a guanosine triphosphate joined to the end nucleotide by a 5′−5′ linkage, rather than the normal 5′−3′ linkage. A methyl group is added to the guanosine, and in some cases to one or both of the ribose residues on the first and second nucleotides (Figure 6.6). Throughout the book we shall use a cartoon cap to depict a cap on the 5′ end of an RNA molecule.
Figure 6.6 Cap on 5’ end of RNA. The inset shows the structure of the cartoon cap, which depicts a methylated guanosine triphosphate linked to the first nucleotide by a 5′−5′ linkage.
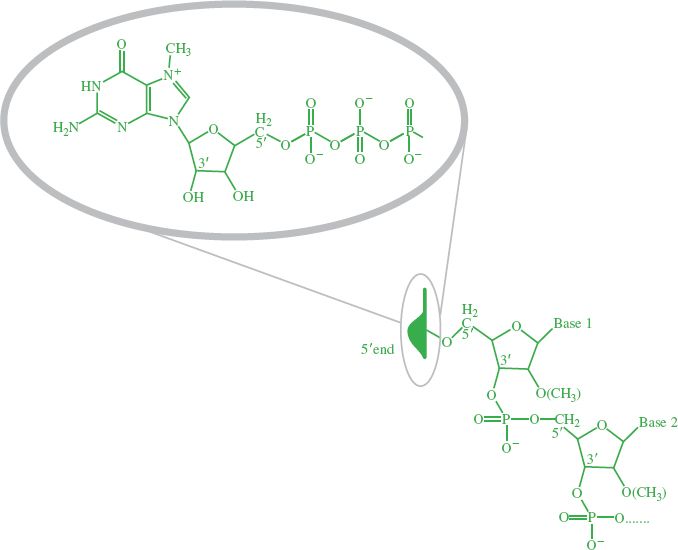
Most eukaryotic cell and viral mRNAs have a cap at their 5′ end. The cap is involved in:
- transport of mRNA from the nucleus to the cytoplasm;
- protection of the mRNA from degradation by exonucleases;
- initiation of translation.
The enzymes that normally cap RNA are listed in Table 6.1. The capping enzymes of the cell are located in the nucleus, and most of the viruses that carry out transcription in the nucleus (Figure 6.5) use these enzymes. Influenza viruses transcribe their genomes in the nucleus, but they do not use the capping enzymes. Instead they “snatch” caps from cell mRNAs; the complex of virus proteins making up the RNA polymerase binds to cellular mRNA, and then an endonuclease in the complex cleaves the RNA, generally 9–15 nucleotides from the 5′ end. The resulting capped oligonucleotide acts as a primer to initiate synthesis of viral mRNA.
Table 6.1 Enzymes involved in capping the 5’ ends of RNAs
| Enzyme | Activity |
| RNA triphosphatase | Removes one of the three phosphate groups from the 5′ end of the RNA |
| Guanylyl transferase | Adds guanosine monophosphate to the diphosphate terminus |
| Methyl transferases | Add methyl group(s) |
Of the viruses that replicate in the cytoplasm, many encode their own capping enzymes (e.g. poxviruses, reoviruses, coronaviruses), while some “snatch” caps from cell mRNAs (e.g. bunyaviruses) and others produce transcripts that are not capped (e.g. picornaviruses). Translation of mRNAs without caps is initiated by a mechanism that is not dependent upon a cap (Section 6.4.1).
6.3.5 Polyadenylation of transcripts
A series of adenosine residues (a polyadenylate tail; poly(A) tail) is added to the 3′ end of most primary transcripts of eukaryotes and their viruses. Polyadenylation probably increases the stability of mRNAs, and the poly(A) tail plays a role in the initiation of translation (Section 6.4.1). These functions can be provided in other ways, however, as some viruses, such as the reoviruses (Section 13.3.2), do not polyadenylate their mRNAs.
In most cases there is a polyadenylation signal (Figure 6.3) about 10–30 bases upstream of the polyadenylation site. The polyadenylation signal AATAAA was first characterized in simian virus 40 in 1981. It has since been found that this sequence is used by many other animal viruses, such as HIV-1 (Section 18.4.3) and Rous sarcoma virus, as well as by animal cells. Some viruses use other sequences as polyadenylation signals; for example, the mammalian hepadnaviruses (Section 19.8.3) use TATAAA, a sequence that can function as a TATA box in other contexts!
In most cases the poly(A) tail is added by the following mechanism. During transcription the RNA polymerase proceeds along the template past the polyadenylation signal and the polyadenylation site. The newly synthesized RNA is then cleaved at the polyadenylation site and the poly(A) tail is added step by step by a complex of proteins, including a poly(A) polymerase. Alternative mechanisms to polyadenylate mRNAs have been evolved by some viruses, including the picornaviruses (Section 14.4.4) and the rhabdoviruses (Section 15.4.2).
6.3.6 Splicing transcripts
Some eukaryotic cell primary transcripts are functional mRNAs, but most contain sequences (introns) that are removed in the nucleus. The remaining sequences (exons) are spliced at specific donor sites and acceptor sites to produce the mRNAs (Figure 6.3). A primary transcript may be cut and spliced in more than one way to produce two or more mRNA species.
Some primary transcripts of viruses that replicate in the nucleus are processed in the same way to produce the virus mRNAs. The first evidence of split genes, as they are known, was reported in 1977 after studies with adenoviruses. Further examples of viruses that have split genes are herpesviruses (Section 11.5.2), parvoviruses (Section 12.4.3) and retroviruses (Section 17.3.4).
The simplest type of split gene consists of two exons separated by one intron, but some are much more complex; gene K15 of Kaposi’s sarcoma-associated herpesvirus has eight exons and seven introns. The HIV-1 genome has a number of splice donor sites and acceptor sites; cutting and splicing the primary transcript results in more than 30 species of mRNA (Section 18.4.3).
6.4 TRANSLATION IN EUKARYOTES
Most viruses synthesize their proteins utilizing the entire translation machinery of the host cell (ribosomes, enzymes, other proteins, and transfer RNAs (tRNAs)). Some large DNA viruses, however, encode their own tRNAs, while the mimivirus (Section 3.2.1) also encodes a number of the enzymes and other protein components of the translation machinery.
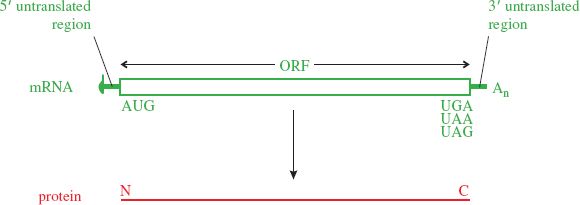
A typical eukaryotic mRNA is monocistronic; that is, it has one ORF from which one protein is translated (Figure 6.7). Sequences upstream and downstream of the ORF are not translated. Some virus mRNAs have large ORFs that encode polyproteins, large proteins that are cleaved to form two or more functional proteins.
Figure 6.7 Translation from a monocistronic mRNA. There is one open reading frame (ORF), usually starting at the first AUG codon from the 5′ end of the mRNA and ending at a stop codon (UGA, UAA, or UAG). Translation occurs in the 5′ to 3′ direction, the N terminus of the protein being synthesized first.
Stay updated, free articles. Join our Telegram channel

Full access? Get Clinical Tree


