Gene
Polymorphism(s)
Drug/drug class
Type of ADR
Comment on evidence
HLA–B
HLA-B*1502
Carbamazepine
Stevens-Johnson syndrome
Recommended in Asian ancestry
CYP2D6
Partially or totally inactive alleles
Pimozide
Arrhythmia
Recommended for doses >4 mg/day in adults and >0.05 mg/kg/day in children
CYP2D6
Partially or totally inactive alleles
Polypharmacy
Overall risk of ADRs
High evidence, probably will be recommended
POLG
A467T, W748S
Valproate
Liver toxicity
Recommended in children/adolescents
CPS1
rs1047891
Hyperammonemia
Recommended in case of suspected urea cycle disorder
HLA-DQB1
6672G>C
Clozapine
Agranulocytosis
Probably will be recommended in the near future
HLA–B
158T
Promising
HTR2C
−759C/T
Antipsychotics
Metabolic ADRs
Promising
MC4R
rs489693
Promising
Leptin
−2548A/G
Promising
CNR1
rs806378, rs1049353
Promising
HTR2A
rs6311, rs6313
Antidepressants
Overall tolerability
Promising
SLC6A4
5-HTTLPR, rs25531
Promising
DRD2
rs1800497
Antipsychotics
Tardive dyskinesia
Promising
HTR2A
102CC, −1438GG
Promising
CYP2D6
Partially or totally inactive alleles
Promising
HSPG2
rs2445142
Parkinsonism
Promising
ZFPM2
rs12678719
Promising
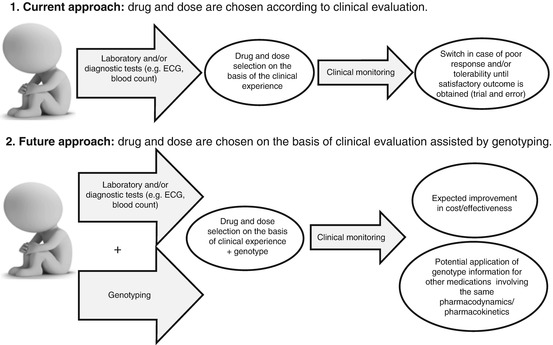
Fig. 7.1
Schematic description of the current approach in the choice of psychotropic medications (1) and expected future approach based on both clinical judgment and genotype information (2)
7.1.2 Pharmacogenetics in Focus
Approximately 0.5 % of the DNA sequence is responsible for phenotype (i.e., somatic) differences among humans. This difference consists in di-, tri-, and tetranucleotide repeats (satellite sequences) and large variants >1 kbp due to deletions, insertions, or duplications (copy number variants, CNV) and nucleotide substitutions. Over three million substitutions distinguish the individual genome, and over 80 % of them are in the form of single-nucleotide substitution polymorphism (SNP). Therefore, it has been estimated that SNPs account for over 80 % of the variability between humans, including liability to ADRs (Roberts et al. 2010). The Human Genome Project (HGP), which started in 1990 and was completed in 2003, with further analysis still being published, has made possible to determine the sequence of chemical base pairs which make up DNA and to identify and map the approximately 20,000–25,000 genes of the human genome from both a physical standpoint and functional standpoint.
The largest part of available data regarding the pharmacogenetics of ADRs has been obtained through candidate gene studies. Candidate genes are selected on the basis of their biological role and polymorphisms on the basis of their functional role (i.e., a known impact on gene function resulting in a variation in the level/function of the product), tagging properties (linkage disequilibrium with near variants), or position (in regulatory regions). In the last decade, genome-wide association studies (GWAS) were introduced and have rapidly expanded, since they provide hundreds of thousands of variants, thanks to the array technology without the need of any a priori hypothesis. Obviously, GWAS have some limitations (in particular the limited covering of genetic polymorphisms provided by the currently available platforms and the common difficulty in explaining the biological meaning of findings).
7.2 Pharmacogenetics of Antidepressant-Induced Side Effects
Antidepressant drugs are among the most frequently prescribed drugs worldwide. Selective serotonin reuptake inhibitors (SSRIs) and serotonin-noradrenaline reuptake inhibitors (SNRIs) are usually better tolerated than first-generation antidepressants (tricyclic antidepressants or TCAs; inhibitors of monoamino oxidase or MAOIs). Nevertheless, some patients require treatment with the latter group of antidepressants or are genetically predisposed to develop idiosyncratic ADRs at therapeutic dosages (an abnormal effect due to polymorphisms in metabolic enzymes or target molecules) or to have an increased risk of overdose from drug with a well-defined and wide therapeutic index. Some patients are at higher risk of severe ADRs because of medical comorbidities, concomitant medications, and/or old age. Finally, the issue of treatment-induced suicidal ideation (TESI) when prescribing antidepressants led regulatory authorities to issue warnings to clinicians (US Food and Drug Administration 2006). Pharmacogenetics can provide clinically useful information on the safe use of antidepressant drugs, especially in patients at increased risk of ADRs due to individual, pharmacological, and/or environmental factors.
7.2.1 Treatment-Emergent Suicidal Behavior (TESI)
Family studies support a genetic contribution to suicidal behavior (SB) (Brent and Mann 2005). Candidate gene studies were focused especially on serotonin-related genes since low central nervous system (CNS) serotonin (5-HT) turnover was demonstrated in SB (Mann 2003). Consistently, the association of SB with variants in the serotonin transporter gene (SLC6A4) (Li and He 2007) and the tryptophan hydroxylase 1 gene (TPH1 that codes for the rate-limiting enzyme responsible for 5-HT biosynthesis) (Bellivier et al. 2004) was supported at meta-analytic level. The noradrenergic system and especially the alpha 2A-adrenergic receptor gene (ADRA2A) have also been implicated in SB (Escriba et al. 2004; Sequeira et al. 2004) with an effect that may be higher in patients treated with noradrenergic antidepressants and in males (Perroud et al. 2009). The noradrenergic system is implicated in the modulation of aggressive and impulsive behaviors, and enhanced noradrenergic activity may have a role in treatment-emergent suicidal behavior (TESI). Thus, the enhanced activity or hypersensitivity of ADRA2A receptors may be associated with higher suicidal ideation during treatment with noradrenergic antidepressants.
Other candidate gene studies suggested the involvement of brain-derived neurotrophic factor gene (BDNF) and the gene coding for its receptor, the neurotrophic tyrosine kinase receptor type 2 (NTRK2) (Perroud et al. 2008). These genes are involved in the regulation and growth of 5-HT neurons and are mediators of neural plasticity in response to acute and chronic stress. The cyclic adenosine monophosphate (cAMP) response element binding (CREB1) protein gene is involved in the regulation of BDNF expression, and it has also been associated with TESI (Perlis et al. 2007).
In addition to dysregulation in the monoaminergic and neurotrophic systems, the hypothalamic-pituitary-adrenal (HPA) axis and inflammatory pathways have been also proposed as modulators of TESI. Indeed, altered sensitivity to glucocorticoids and increase in proinflammatory cytokines were demonstrated in depression and particularly in suicide victims. BDNF and 5-HT positively modulate neurogenesis in the hippocampus (a pivotal area involved in depression pathogenesis), and the hippocampus in turn regulates HPA axis function and response to stress. In depressed subjects, hippocampal atrophy (due to reduced cell proliferation, cell survival, and cell differentiation) promotes impaired regulation of HPA axis activity (Mahar et al. 2014). FK506-binding protein 5 (FKBP5) gene, which codes for a protein that decreases the sensitivity of the glucocorticoid receptor to the effect of corticosteroids, was suggested as a modulator of TESI (Mandelli and Serretti 2013).
The role of glutamate in modulating mood and antidepressant response has been increasingly recognized, due to observations that existing antidepressants modulate various glutamatergic pathways. Disrupted glutamatergic-noradrenergic interactions at the level of the stress-sensitive locus coeruleus (LC) were demonstrated in depression and suicide victims (Chandley et al. 2014). Accordingly, polymorphisms in glutamate receptor genes GRIK2 and GRIA3 were associated with TESI during SSRI treatment (Laje et al. 2007).
Genome-wide association studies (GWAS) did not report the above genes among their top findings but outlined other genes that may involved in TESI. Associations were found for genetic variants within the loci encoding papilin (PAPLN) and IL-28 α-receptor (IL28RA) genes (Laje et al. 2009) and in the vicinity of the guanine deaminase (GDA) gene (Perroud et al. 2012). IL28RA encodes a cytokine receptor, while papilin is involved in the regulation of extracellular matrix remodeling, a process that affects the release of bioactive fragments that function as immune modulators (Korpos et al. 2009). The involvement of these genes in TESI is therefore consistent with the inflammation theory of depression and SB. On the other hand, GDA encodes an enzyme responsible for the hydrolytic deamination of guanine and is probably involved in microtubule assembly. No clear biological rationale links this gene to TESI. A recent development of GWAS is based on multimarker analyses given the hypothesis that multiple genetic variants contribute to complex phenotypes such as TESI. A cluster of 79 SNPs demonstrated a 94 % probability of predicting the nonoccurrence of TESI (negative predictive value), even with only a 48 % probability of correctly identifying TESI (positive predictive value) (Menke et al. 2012), but with no confirmation of this finding so far.
Given the complex and multifactorial pathogenesis of TESI, it seems unlikely that genotyping could be able to prevent it adequately.
7.2.2 Cardiovascular Side Effects
Tricyclic antidepressants (TCAs) have a higher risk of cardiovascular side effects compared to other antidepressants, even in patients with no previous cardiovascular disease (Pacher and Kecskemeti 2004). The most common among such side effects is the slowing of intraventricular conduction, manifested by prolonged PR, QRS, and QT intervals on the standard ECG, and orthostatic hypotension. TCAs have been demonstrated to exert I/A class antiarrhythmic effects and antinoradrenergic and anticholinergic effects that are responsible for their cardiovascular ADRs.
The identification of genetic polymorphisms predicting the risk of intraventricular conduction alterations induced by antidepressants could help clinicians to prevent life-threatening side effects. So far, a number of genes have been associated with arrhythmia: SCN5A, SCN4B, CACNL1AC, KCNH2, KCNQ1, KCNE1, ANK2, ALG10, KCNJ2, KCNE2, RYR2, KCND3, KCND2, ACE, NOS1AP, CASQ2, and Rad (Drago et al. 2008). These genes are good candidates for the definition of a genetic proarrhythmic profile, but evidence is still lacking for antidepressant drugs.
Changes in systemic blood pressure are possible especially during treatment with antidepressants that affect the activity of the noradrenergic system (norepinephrine (NE) reuptake, transport, and elimination from the synapse). Blood pressure changes are mediated through the autonomic nervous system in part by the neurotransmitter NE. Selective NE transporter (NET) blockade creates a phenotype that resembles idiopathic orthostatic intolerance (Schroeder et al. 2002), but the gene encoding this transporter (SLC6A2) was very marginally associated with blood pressure changes during treatment with duloxetine (an SNRI antidepressant drug). Other noradrenergic genes (ADRB2, coding for the adrenergic beta 2 receptor, and COMT, encoding the main enzyme involved in NE metabolism) do not apparently play a role in the risk of blood pressure changes during treatment with duloxetine (Fijal et al. 2013). In addition, 5-HT binding to the serotonin-2A receptor has been associated with vasoconstriction and hypertension, and HTR2A gene has consistently been associated with essential hypertension in women (Liolitsa et al. 2001). Nevertheless, evidence suggesting an effect of this gene on blood pressure increase during treatment with duloxetine appears limited (Fijal et al. 2013).
The SSRIs fluoxetine and paroxetine demonstrated a relatively high receptor affinity for adrenergic beta receptors in vitro, suggesting an increased propensity of affecting cardiovascular parameters compared to other SSRIs. Interestingly, a polymorphism in the gene encoding the adrenergic beta 1 receptor (ADRB1) was found to modulate blood pressure and heart rate values in patients treated with these antidepressants (Thomas et al. 2010), but the potential clinical impact of this appears to be limited.
Finally, a group of clinically relevant side effects affecting the cardiovascular system are hemorrhagic complications, whose risk is increased during treatment with SSRI antidepressants, especially in some at-risk conditions (e.g., concomitant treatment with anticoagulant or antiaggregant drugs and surgery). 5-HT is a strong vasoconstrictor and a relatively weak platelet activator. At rest, 5-HT is stored in platelets, but after platelet activation, it is released into the circulation, together with other aggregating factors such as adenosine diphosphate (ADP) and adrenaline, and it acts as a stimulus for platelet aggregation. The 5-HT transporter is present on platelet surface and is necessary to transport 5-HT into the platelet, since platelets themselves do not produce 5-HT but are dependent on its uptake from the blood. Thus, blockade of the 5-HT transporter with an SSRI leads to a lower concentration of 5-HT in the platelet. Given this mechanisms, SLC6A4 gene (encoding the 5-HT transporter) appears to be an optimal candidate for affecting the risk of bleeding during SSRI treatment. An insertion/deletion polymorphism (named 5-HTTLPR) in the promoter region of SLC6A4 showing two variant alleles (a short (S) allele and a long (L) allele) has been particularly studied since the S variant is associated with a nearly 50 % reduction in basal expression of the 5-HT transporter (Heils et al. 1996). Despite the interesting functional impact of this polymorphism, the available knowledge does not suggest any major impact of this variant on platelet function (Abdelmalik et al. 2008; Hougardy et al. 2008).
In conclusion, no genetic variants are known to significantly affect the risk of cardiovascular side effects during antidepressant treatment, but very few studies were focused on this topic. The identification of polymorphisms affecting the risk of proarrhythmic effects appears to be the most relevant issue under the clinical point of view.
7.2.3 Weight Gain, Sexual Dysfunction, and Other Side Effects
Sexual dysfunction, weight gain, and gastrointestinal side effects (dry mouth, constipation, diarrhea, and nausea) are the most frequent side effects induced by antidepressant drugs and thus the most frequently responsible for early treatment discontinuation. On the other hand, antidepressant-related hyponatremia is a relatively rare but potentially fatal antidepressant-induced ADR.
Some polymorphisms (5-HTTLPR, rs25531, intron 2 VNTR, or STin2) of the 5-HT transporter gene (SLC6A4) have been repeatedly investigated for association with the overall risk of antidepressant-induced side effects. Available evidence mainly suggests that carriers of the 5-HTTLPR/rs25531 short alleles show lower treatment tolerability while STin2 probably does not exert a significant influence on this phenotype (Garfield et al. 2014; Fabbri et al. 2013). HTR1A rs6295 and HTR2A rs6311/rs6313 are also promising variants that contribute to individual antidepressant tolerability (Fabbri et al. 2013; Garfield et al. 2014).
Weight gain is a quite common side effect of treatment with antidepressant drugs. Genes pertaining to the serotonergic system have been particularly investigated in relation to this ADR, since 5-HT has been implicated in the control of eating behavior and body weight by hypothalamic serotonergic receptor mechanisms (De Vry and Schreiber 2000). HTR2C gene (coding for 5-HT2C receptor) was proposed as a modulator of the risk of weight gain during both antidepressant and antipsychotic therapies (Altar et al. 2013). The stimulation of hypothalamic 5-HT2C receptors leads to a behaviorally specific hypophagic effect by accelerating satiety processes (De Vry and Schreiber 2000), providing a biological rationale that supports the pharmacogenetic finding.
Catechol-O-methyltransferase (COMT) and tryptophan hydroxylase type I (TPH1) encode pivotal enzymes in the catabolism and synthesis of 5-HT, respectively. COMT rs4680 and TPH1 rs18532 were demonstrated to modulate the risk of weight gain during antidepressant treatment independently from age and gender (Secher et al. 2009). GNB3 (that encodes the β3 subunit of the G protein complex) is involved in the downstream signaling cascade following monoamine receptor activation. A functional polymorphism in this gene (C825T) was associated with the improvement of neurovegetative symptoms of depression during treatment, and the TT genotype was found to be a predictor of greater weight gain during treatment with nortriptyline (a drug of the TCA class) (Keers et al. 2011).
ADRA2A gene (encoding adrenergic α2 receptor) is a primary target of mirtazapine, a noradrenergic and specific serotonergic antidepressant (NaSSA). Mirtazapine is more likely to cause weight gain or increased appetite compared to SSRIs or SNRIs (Watanabe et al. 2011; Lee et al. 2009); thus the effect of ADRA2A-1291C/G polymorphism on the risk of weight gain during treatment with this antidepressant should be investigated further.
Sexual dysfunction in patients with major depression may be triggered or exacerbated by treatment with antidepressants (especially SSRIs, TCAs, and venlafaxine), with a prevalence of up to 50–70 % (Fava and Rankin 2002). Serotonergic, dopaminergic, noradrenergic, and glutamatergic systems are those primarily hypothesized to be involved in the pathogenesis of this ADR. In general, reduction of 5-HT function facilitates, whereas enhancement inhibits, sexual behavior, with the 5-HT transporter, 5-HT1A, 5-HT1B, 5-HT2A/B, and 5-HT7 receptors being the primary molecular players involved (Olivier et al. 2011). Furthermore, the blockade of 5-HT transporters may decrease the concentrations of dopamine and noradrenaline in the mesolimbic system by activating serotonin 5-HT2C receptors (Strohmaier et al. 2011); these two neurotransmitters play a role in the modulation of sexual arousal and sexual motivation. Indeed, dopamine antagonists inhibit copulation among male rats, whereas agonists have the opposite (facilitating) effect. Similar effects were demonstrated also for glutamate antagonists and agonists, respectively (Dominguez et al. 2006; Dominguez and Hull 2005).
The promoter polymorphism 5-HTTLPR rs25531 of the 5-HT transporter gene (SLC6A4) has been investigated as predictor of antidepressant-induced sexual dysfunction. Results suggested a higher risk in high-expressing genotypes (long alleles) of the polymorphism (Garfield et al. 2014), and the effect appears to be dependent on age (Strohmaier et al. 2011). HTR1A and HTR2A genes may also contribute to the development of this ADR (Garfield et al. 2014; Bishop et al. 2006).
Consistently with the involvement of glutamate in the modulation of sexual behavior, in SSRI-treated patients, multiple genes encoding glutamatergic receptors were associated with a decrease in libido (GRIA3 and GRIK2), difficulty achieving orgasm (GRIA1), and difficulty developing an erection (GRIN3A) (Perlis et al. 2009). Currently, no data are available on the contribution of polymorphisms in noradrenergic and dopaminergic genes to the risk of antidepressant-induced sexual dysfunction.
Gastrointestinal ADRs are sometimes disabling side effects that usually emerge in the initial phases of antidepressant treatment. Variants in genes belonging to the serotonergic system were the most studied in relation to these ADRs, since 80 % of the body 5-HT stores are located in enterochromaffin cells of the gut and serotonin plays a central role in the regulation of motility and secreting activity of the gastrointestinal tract. HTR3B gene was associated with paroxetine-induced gastrointestinal side effects (Tanaka et al. 2008; Sugai et al. 2006), and HTR2A was shown to exert a synergistic effect with CYP2D6 gene polymorphisms in the prediction of fluvoxamine-induced gastrointestinal side effects (Suzuki et al. 2006). CYP2D6 gene was associated also with venlafaxine (a SNRI)-induced gastrointestinal ADRs (Shams et al. 2006) and with broad antidepressant-induced ADRs (Rau et al. 2004). Polymorphisms in genes encoding cytochrome P450 enzymes (CYP450) are responsible for variations in drug metabolizing rapidity, including antidepressant drugs. Consequently, genetic variations in CYP450 genes could influence the occurrence of several ADRs, including the risk of overdose; this issue is discussed in the section “Toxicity from Overdose.” P-glycoprotein (P-gp), an ATP-driven efflux pump that regulates the uptake of drugs through organ barriers, is another protein influencing the pharmacokinetics of antidepressants through the regulation of drug distribution in the body. Polymorphisms in the gene coding for this transporting protein (ABCB1) may influence gastrointestinal complaints and sexual side effects during antidepressant therapy (de Klerk et al. 2013).
Hyponatremia is a potentially fatal side effect of antidepressant drugs. It occurs in one in 200 elderly patients per year receiving fluoxetine and paroxetine, two commonly used SSRIs (Wilkinson et al. 1999). There is little pharmacogenetic evidence with regard to this ADR, but lower mean serum sodium concentrations were shown to be present in CYP2D6 poor metabolizers (PMs) in comparison with CYP2D6 extensive metabolizers (EMs). As a result, CYP2D6 PMs might be at increased risk of developing hyponatremia (Kwadijk-de Gijsel et al. 2009).
GWAS did not outlined particularly interesting findings for any of the above ADRs, but different genes were proposed as putative candidates compared to candidate gene studies. SACM1L (coding for the phosphatidylinositide phosphatase SAC1) gene was associated with bupropion-induced sexual dysfunction, even though this antidepressant is not commonly responsible for this ADR, resulting in limited clinical utility. SAC1 is an integral membrane protein of the endoplasmic reticulum and the Golgi apparatus that plays a direct role in growth factor signaling; thus alterations in the activity of this enzyme may lead to disruptions in the cellular secretory machinery and hormone-neurotransmitter secretion, with possible consequences on sexual functioning (Clark et al. 2012). A later extension of the study by Clark et al. suggested that EMID2 (EMI domain containing 2) gene may affect SSRI-induced vision/hearing side effects, LAMA1 (laminin, alpha 1) gene and the rs16965962 SNP (in a gene desert on chromosome 7) may influence overall SSRI tolerability, while AOX2P gene may be related with dizziness. EMID2 encodes the protein collagen α-1 chain that is involved in the regulation of corneal collagen fibrillogenesis (Rada et al. 1993). On the other hand, for LAMA1 and AOX2P, it is not so easy to hypothesize a biological rationale explaining the reported GWAS findings.
7.2.4 Toxicity from Overdose
The risk of overdose from antidepressant drugs was reduced significantly after the introduction of SSRIs and SNRIs compared to earlier antidepressants (TCAs and MAOIs). Despite the reduction of life-threatening reactions, 5-HT toxicity can still result from serotonin excess in the CNS from serotonergic drugs. Serotonin syndrome is a potentially life-threatening condition characterized by myoclonus, hyperreflexia, sweating, shivering, incoordination, and mental status changes. Furthermore, the risk of cardiac arrhythmia is another potentially fatal manifestation following overdose especially from TCAs but possibly also from venlafaxine and IMAOs.
The genetic variants that were hypothesized to influence the risk and severity of overdose symptoms are mainly those in genes coding for CYP450 enzymes and P-glycoprotein (P-gp) gene (ABCB1), the products of which are involved in the distribution and metabolism of antidepressant drugs.
CYP2D6 and CYP2C19 encode the P450 isoenzymes that are mostly involved in antidepressant metabolism. The level of CYP enzyme activity is dependent on genetic polymorphisms and allows the distinction of different metabolizing groups. The wild-type genotype results in extensive metabolizers (EM), while the intermediate metabolizer (IM) is characterized by the presence of one wild-type allele plus a partially or totally defective allele. Poor metabolizers (PMs) have a combination of two partially or totally defective alleles, and the ultrarapid metabolizer (UM) category exists only for CYP2D6 and is usually due to multiple copies of normal alleles.
The available evidence suggests that CYP2D6 PMs have lower tolerance to TCAs as well as to venlafaxine (a SNRI drug), whereas they have an average tolerance to other antidepressants. Based on literature, CYP2D6 PMs are expected to have a concentration-to-dose ratio (C:D ratio) of 4–6 for TCAs, whereas CYP2D6 EMs are expected to have a C:D ratio of 0.5–1.5. Dose adjustments for different metabolizing groups were calculated, even if prospective validations should be performed before routine clinical application (Porcelli et al. 2011). Polypharmacy is likely to represent a valid indication for CYP2D6 genotyping to minimize the risk of toxicity from drug-drug interactions in PMs (Laje 2013).
Finally, P-gp limits drug uptake into key organs such as the brain, and fatal intoxication from venlafaxine overdose was associated with C1236T and C3435T polymorphisms in ABCB1 gene (Karlsson et al. 2013).
7.3 Pharmacogenetics of Antipsychotic-Induced Side Effects
Antipsychotic drugs are the mainstay of treatment for schizophrenia and related disorder and have improved schizophrenia prognosis significantly since their introduction in the 1950s. In recent years, the use of second-generation antipsychotics (SGAPs) has been indicated also for the treatment of several phases of bipolar disorder or augmentation in major depressive disorder with benefits being seen especially in more severe cases. Both first-generation antipsychotics (FGAPs) and SGAPs carry the risk of severe and sometimes debilitating ADRs, whose clinical relevance has been increased in proportion with the expansion of their clinical use in terms of increasing indications and treatment duration.
7.3.1 Antipsychotic-Induced Weight Gain
Weight gain is a major health problem encountered during treatment with antipsychotics especially SGAPs due to a high risk of obesity and other metabolic conditions (Dickerson et al. 2006). Weight gain is associated with significant variability among individuals, and genetic factors play an important role, estimated to be around 60–80 % through twin and family studies (Gebhardt et al. 2010). Given that cardiovascular disease is the primary cause of excess of mortality among severe psychiatric diseases (Osborn et al. 2007), the identification of genetic predictors of antipsychotic metabolic ADRs could be a turning point in the treatment of schizophrenia and bipolar disorder.
Several pharmacogenetic studies have investigated the genes that could influence antipsychotic-induced weight gain (AIWG), with focus mainly on homeostatic regulators expressed in hypothalamic areas that belong to the complex network that regulates appetite and satiety.
The most replicated pharmacogenetic association is the serotonergic receptor 5-HTC2 (HTR2C) gene that is responsible for 5-HT central anorexigenic action on the hypothalamic nuclei. Consistently, antagonists of 5-HTC2 receptors, such as clozapine and olanzapine, promote appetite increase (Bonhaus et al. 1997). Carriers of the minor T allele of the promoter polymorphism −759C/T appear to be protected from substantial gain in weight (Sicard et al. 2010).
Leptin and melanocortin receptor 4 (MC4R) are other essential components of one of the most important hypothalamic satiety signals. Leptin is mainly synthesized in the adipocytes of white adipose tissue and activates leptin receptors in the arcuate nucleus of the hypothalamus, resulting in a feeling of satiety. The leptin-2548A/G polymorphism may interact with the HTR2C −759C/T variant in affecting AIWG (Reynolds 2012). Neurons of the arcuate nucleus also express MC4R, activation of which decreases food intake while elevating energy utilization (Fani et al. 2014). This gene has been consistently associated with AIWG by four independent studies (Shams and Muller 2014).
Cannabinoid receptor 1 (CNR1) gene and the fatty acid amide hydrolase (FAAH) gene have been suggested as genetic factors involved in AIWG (Shams and Muller 2014), consistently with the observation that they play an important role in the mediation of leptin anorexigenic action. Some SNPs within CNR1 in particular provided encouraging findings.
Other candidate genes provided less convincing evidence for an association with AIWG, among them being ghrelin (GHRL) and neuropeptide Y (NPY), which act as antagonists of the leptin-induced satiety signal, other hypothalamic neuroendocrine regulators (FTO and PMCH genes), and histamine receptor 1 (HRH1 gene, consistently to the observation that central histaminergic transmission contributes to the modulation of the motivational aspect of appetite and physical activity (Torrealba et al. 2012)). The T allele of GNB3C825T polymorphism was considered to be particularly interesting since it is associated with a G protein β3 splice variant and previously described associated with obesity in several ethnic groups. Nevertheless, the available evidence mainly does not support the involvement of this variant in AIWG (Souza et al. 2008). Furthermore, polymorphisms in AMP-activated protein kinase (AMPK gene, a central molecule integrating nutrient and hormonal signals to maintain energy homeostasis), relaxin-3 (RLN3, a member of the insulin/relaxin pathway) and its receptors (RXFP3 and RXFP4), tumor necrosis factor-α (TNF–a, a proinflammatory cytokine), and methylenetetrahydrofolate reductase (encoded by MTHFR, involved in nuclear methylation and gene expression regulation) were suggested to modulate AIWG (Muller et al. 2013; Kao and Muller 2013).
A GWAS was aimed to identify genetic risk factors for metabolic side effects in patients treated with psychopharmacological medications (Athanasiu et al. 2012). SNP rs7838490 (8q21.3 region) was associated with BMI alterations, while rs11615724 (12q21) was associated with the effect of medications on decreasing HDL-C levels. Both markers are in intergenic regions. rs7838490 is located upstream of the gene matrix metalloproteinase 16 (MMP16), and it may regulate the expression of MMP16 and affect tumor necrosis factor receptor superfamily, member 1A (sTNFRSF1A), which may be involved in lipid regulation.
7.3.2 Antipsychotic-Induced Extrapyramidal Side Effects
The greater part of pharmacogenetic studies focused on genetic risk factors of antipsychotic-induced tardive dyskinesia, the most severe among extrapyramidal side effects (EPS) due to its tendency to persist over time, its treatment resistance, and its high frequency (around 20 % of patients after prolonged treatment (Kane et al. 1988)). Genes are hypothesized to be a relevant factor in the risk of tardive dyskinesia, as suggested by increased risk of tardive dyskinesia in affected families (Muller et al. 2013). Genes that influence the pharmacokinetics, pharmacodynamics, and oxidative stress associated with antipsychotics have been studied for tardive dyskinesia risk. In particular, the gene coding for the dopamine receptor 2 (DRD2), one of the main targets of antipsychotics (especially for those drugs with a higher risk of inducing this ADR), was suggested to be involved in the risk of this EPS by various independent studies (especially the Taq1A polymorphism or rs1800497). The minor (T) Taq1A allele has been associated with a 40 % reduction in striatal D2 receptor density (according to in vitro assays and in vivo imaging studies); this allele appears to be protective against tardive dyskinesia (Lencz and Malhotra 2009). DRD2 may also be implicated in the risk of akathisia, even though only preliminary evidence is available (Muller et al. 2013). A functional missense mutation in dopamine 3 receptor (DRD3, another target of antipsychotics) gene (ser9gly or rs6280) has been suggested to modulate the risk of tardive dyskinesia, but meta-analytic results did not support this hypothesis (Tsai et al. 2010). 5-HT2A receptor (HTR2A) gene is a target of atypical antipsychotics, and it has been implicated in their reduced extrapyramidal side effect profile (Meltzer 2012). HTR2A gene has also been associated with tardive dyskinesia susceptibility by several candidate gene studies (Segman et al. 2001; Gunes et al. 2007; Wilffert et al. 2009).
The highly polymorphic gene CYP2D6 is responsible for the hepatic metabolism of several commonly prescribed antipsychotics. CYP2D6 poor metabolizer status (homozygosity for null alleles) or intermediate metabolizer status (null allele heterozygosity) were associated with 1.64- and 1.43-fold greater odds of developing tardive dyskinesia on the basis of literature meta-analysis (Patsopoulos et al. 2005).
Other genes were investigated in relation to antipsychotic-induced EPS, among which were the glutamate receptor GRIN2A, BDNF, TNF–a (Muller et al. 2013), and SOD2 (Lencz and Malhotra 2009). SOD2 encodes for manganese superoxide dismutase, a mitochondrial enzyme involved in oxidative metabolism. SOD2 and TNF–a may be involved in the risk of tardive dyskinesia through neuron oxidative stress.
Stay updated, free articles. Join our Telegram channel

Full access? Get Clinical Tree


