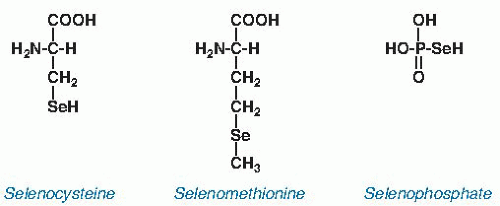
and is considerably less reactive than selenocysteine. It is not known to have a biochemical function distinct from that of methionine.
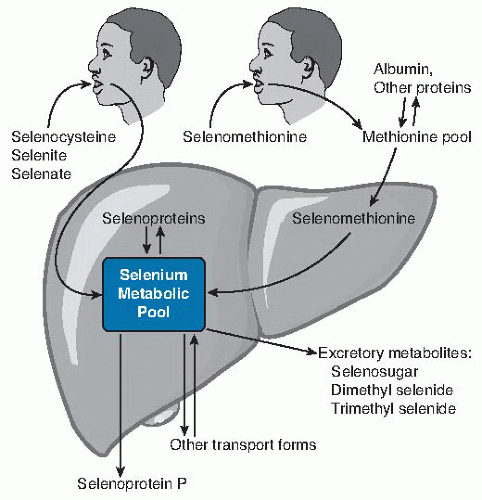 Fig. 14.2. Relationship of dietary form of selenium with tissue forms of the element. Ingested selenocysteine and inorganic selenium forms selenite and selenate enter the selenium metabolic pool directly (more detail in Fig. 14.3) directly. Selenomethionine enters the methionine pool and is incorporated into methionine-containing proteins throughout the body. When selenomethionine is metabolized to selenocysteine by the transsulfuration pathway, it enters the selenium metabolic pool. In liver, the selenium metabolic pool produces hepatic selenoproteins and selenoprotein P (SEPP1) for export. Homeostasis of selenium is maintained by the production of excretory metabolites and transport forms of selenium. |
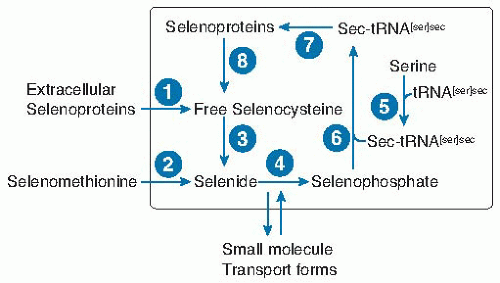 Fig. 14.3. Selenium metabolic pool in a typical nonliver or kidney cell. Selenium enters the cell as selenocysteine from extracellular selenoproteins (probably mainly selenoprotein P) (1) or breakdown of selenomethionine (2) or from unidentified small molecule forms. Free selenocysteine is produced by catabolism of (8) cellular selenoproteins or (1) extracellular selenoproteins. Free selenocysteine does not accumulate because it is metabolized by (3) selenocysteine β-lyase. The resulting selenide is transformed into selenophosphate by (4) selenophosphate synthetase, by using the cosubstrate adenosine triphosphate. The amino acid serine is acylated (5) to the tRNA[ser]sec to form ser-tRNA[ser]sec. In bacteria, selenophosphate is a substrate for selenocysteine synthetase (6), which forms sec-tRNA[ser]sec directly, whereas in eukaryotes a kinase in an additional step phosphorylates serine on the ser-tRNA[ser]sec, which then with selenophosphate is the substrate for selenocysteine synthetase (6). sec-tRNA[ser]sec donates selenocysteine to the growing peptide chain in the synthesis of selenoproteins (7) (see Fig. 14.4B). |
urinary excretion is the primary means whereby body selenium is regulated (14).
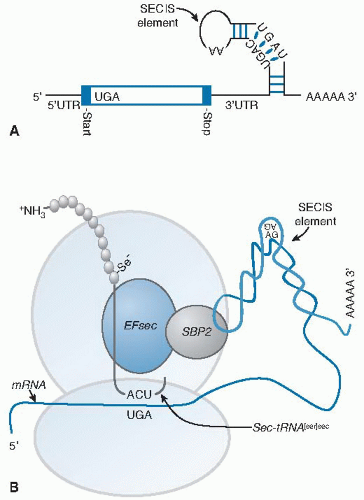 Fig. 14.4. Selenoprotein synthesis. A. The mRNA of a selenoprotein has a UGA in the open reading frame specifying selenocysteine (Sec) incorporation and a specialized stem-loop structure, known as an Sec insertion sequence (SECIS) element, in the 3′ untranslated (3′UTR) region. B. Two trans-acting proteins, SBP2 (SECIS-binding protein 2) and EFsec (elongation factor for selenocysteine), facilitate the recognition of the sec-tRNA[ser]sec by the UGA. SBP2 binds to the SECIS element and interacts with EFsec that has the sec-tRNA[ser]sec attached. This complex delivers the sec-tRNA[ser]sec to the ribosome for incorporation of Sec into the growing polypeptide chain (see Fig. 14.3, step 7). |
TABLE 14.1 SELENOPROTEINS IN HUMANS | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Stay updated, free articles. Join our Telegram channel

Full access? Get Clinical Tree