CHAPTER 42 S Phase and DNA Replication
Accurate replication of DNA, which is crucial for cellular propagation and survival, occurs during the S phase (DNA synthesis phase) of the cell cycle. This chapter begins with a brief primer on the events of replication and then discusses its regulation. Next, the chapter covers the proteins that bind origins of replication and ensure that each region of DNA is replicated once and only once per cell cycle. It closes by discussing how the structure of the nucleus influences replication.
DNA Replication: A Primer
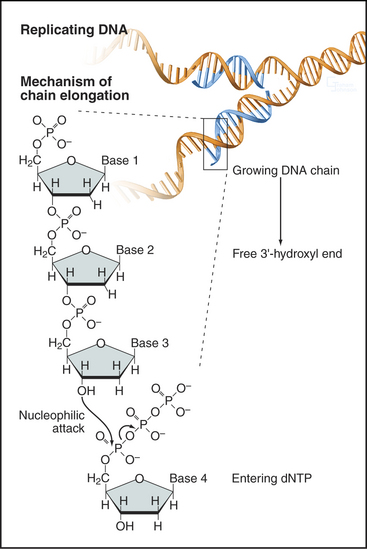
In the basic reaction of DNA replication, the 3′ hydroxyl at the end of the growing DNA strand makes a nucleophilic attack on the α-phosphate of the incoming nucleoside triphosphate to form a phosphodiester bond. This incorporates the nucleotide into the growing chain and releases pyrophosphate (Fig. 42-1). Subsequent hydrolysis of the pyrophosphate provides the driving force for the reaction. This reaction requires the presence of a template strand of DNA that specifies, through base pairing, which of the four nucleoside triphosphates is added to the growing complementary strand.
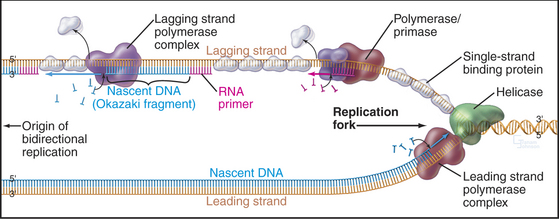
Before discussing DNA replication and its regulation, an introduction to some terminology describing the geometry of replicating DNA is required. The exact site on the chromosomal DNA where replication begins is termed the origin of bidirectional replication. As the termbidirectional implies, two sets of DNA replication machinery head off in opposite directions from the origin. Each set of replication machinery, together with the DNA that it is replicating, is called a replication fork because at the site of replication, one parental DNA molecule splits into two (Fig. 42-2). It is not known whether replication forks move along the DNA like trains along a track or whether the fork sits at a stationary site (referred to as a replication factory) through which the DNA is “reeled in” as it is replicated.
The bidirectional nature of DNA replication causes a fundamental problem, as DNA synthesis invariably proceeds in a 5′ to 3′ direction. Replication of the so-called leading strand poses no problems. This is the strand along which the fork moves in a 3′ to 4′ direction, so the newly synthesized DNA is laid down smoothly in a 5′ to 3′ direction (Fig. 42-2). However, the other template strand faces in the opposite direction, apparently requiring DNA polymerase to synthesize DNA in the wrong direction as the replication fork progresses away from the origin (i.e., adding nucleotides in a 3′ to 4′ direction). No DNA polymerase with this polarity has been found. Instead, this lagging strand replicates in a series of short segments. Every time the DNA strands have been peeled apart (unwound) by 250 nucleotides or so, a polymerase/primase complex (see Fig. 42-11) initiates DNA synthesis on the lagging strand, with the polymerase running back toward the replication origin in a 5′ to 3′ direction. Locally, synthesis on the lagging strand proceeds in a direction opposite to the overall direction of fork movement. Synthesis of each lagging strand fragment stops when DNA polymerase runs into the 5′ end of the previous fragment. Thus, the lagging strand is copied in a highly discontinuous fashion into short fragments known as Okazaki fragments (named after their discoverer [Fig. 42-2]). Fig. 42-11 describes the enzymes and events at the replication fork in greater detail.
Origins of Replication
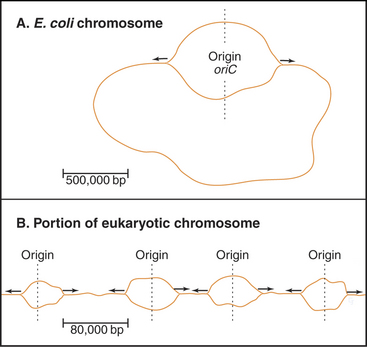
Bacteria such as Escherichia coli replicate their circular chromosomes using two replication forks starting from a single origin of replication (Fig. 42-3A), but eukaryotes must use multiple origins of replication to duplicate their large genomes during a relatively short S phase, which can be limited to as little as a few minutes in some early embryos. These numerous origins are distributed along the chromosome: up to 400 in budding yeast and about 60,000 in human cells. These origins are positioned so that all of the DNA is replicated in the available time, and to be on the safe side, more origins are prepared than are actually needed.
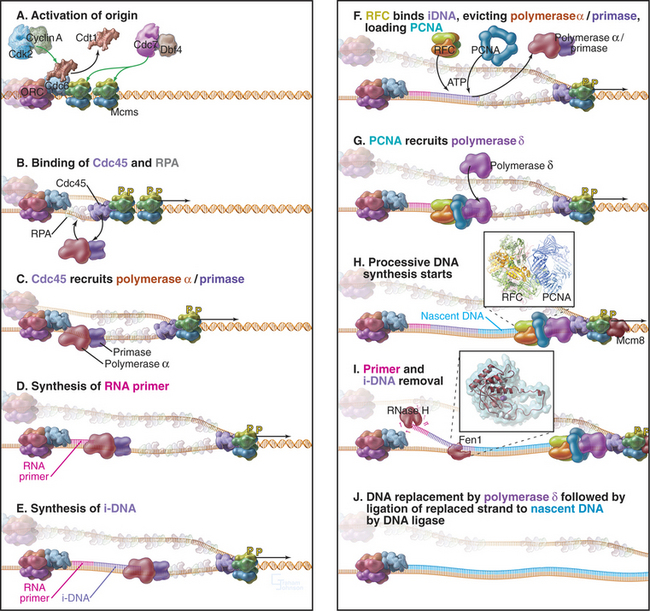
The existence of multiple origins creates a potential hazard: If any origin were used more or less than once per cell cycle, genes would be duplicated or lost. How is the “firing” of all of these origins orchestrated so that each is used once and only once per S phase? Cells manage this problem by a mechanism termed licensing, which ensures that each origin is used once and only once per S phase. Each origin is licensed to replicate once and only once per cell cycle. Replication of the origin removes the license, which cannot normally be renewed until the cell has completely traversed the cycle and has passed through mitosis.
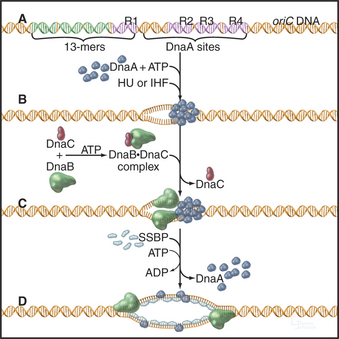
A unit of chromosomal DNA whose replication is initiated at a single origin is termed a replicon. The origin is defined genetically as a replicator element. The classic replicon is the E. coli chromosome (which is 4 × 106 base pairs [bp] in size); this has a single replicator site called oriC (Fig. 42-3). An initiator protein (product of the E. coli DnaA gene [Fig. 42-12]) binds to this origin and either directly or indirectly promotes melting of the DNA duplex, giving the replication machinery access to two single strands of DNA. Other factors bind to the initiator, and their concerted action produces a wave of DNA replication proceeding outward in both directions along the DNA (a replication “bubble”) at about 750 to 1250 bases per second.
An average human chromosome contains about 150 × 106 bp of DNA. Because the replication machinery in mammals moves only about 20 to 100 bases per second (probably reflecting the fact that the DNA is packaged into chromatin [see Chapter 13]), it would take up to 2000 hours to replicate this length of DNA from a single origin. In most human cells, the duration of the S phase is about eight hours. This means that at least 25 to 125 origins of replication would be required to replicate an average chromosome in the allotted time. In fact, origins of replication are much more closely spaced than this. It has been estimated that mammalian origins of replication are spaced about 100,000 to 150,000 bp apart. Thus, approximately 60,000 origins of replication participate in replication of the entire human genome.
Replication Origins in S. Cerevisiae
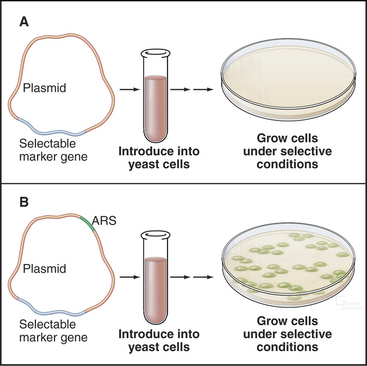
About 400 origins of replication participate in replicating the budding yeast genome. A major breakthrough in understanding DNA replication in S. cerevisiae was the identification of short (100 to 150 bp) segments of DNA that act as replication origins in vivo when cloned into a yeast plasmid (circular DNA molecule). These autonomously replicating sequences (or ARS elements) allow yeast plasmids to replicate in parallel with the cellular chromosomes (Fig. 42-4). ARS elements are often, although not always, bona fide replication origins in their native chromosomal context. Replication always initiates within ARS elements, but not all ARS ele-ments act as origins of DNA replication in every cell cycle.
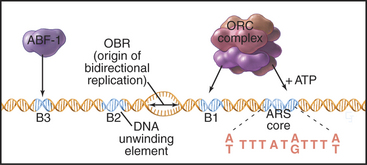
Budding yeast ARS elements share a common DNA sequence motif called the ARS core consensus sequence: 5′-(A/T)TTTAT(A/G)TTT(A/T)-3′ (Fig. 42-5). Single base mutations at several locations within this sequence completely inactivate ARS activity. Other, less well-conserved DNA sequences also contribute to the activity of the ARS as a replication origin. One of these, termed B1, together with the ARS core, forms the binding site for a complex of six proteins (five of which are AAA ATPases) termed the origin recognition complex (ORC [see later section]). The DNA unwinding element is thought to be another short sequence (B2) located a bit further along the DNA. DNA synthesis begins at an origin of bidirectional replication midway between the ORC binding site and the DNA unwinding element.
ORC was identified by its ability to bind the 11-bp ARS core sequence (Fig. 42-5). This binding has two noteworthy features. First, it requires adenosine triphosphate (ATP), which remains associated with the ORC complex. Second, in yeast, the ORC complex remains bound to the origins of replication across the entire cell cycle. Thus, something other than the presence of ORC must be responsible for regulating the periodic activation of origins in the S phase (see Fig. 42-14). In metazoans, ORC behavior is more complex; the largest subunit, Orc1, cycles on and off the DNA in a cell-cycle-regulated manner.
ARS elements typically contain binding sites for other sequence-specific DNA binding proteins, such as transcription factors. For example, a transcription factor called ARS-binding factor 1 (ABF-1) binds to the B3 sequence within the ARS1 element (Fig. 42-5). Deletion of the ABF-1 binding site only slightly reduces the ability of ARS1 to act as a replication origin in vivo. Furthermore, substitution of DNA binding sequences for other transcription factors within the B3 sequence has little effect on replication efficiency.
In addition to their role in DNA replication, several ORC components also seem to regulate heterochromatin formation and transcription (see Chapters 13 and 15). This cross talk between the machinery used for transcription and DNA replication may explain why regions of chromosomes with actively transcribed genes typically replicate early in the S phase (see the discussion that follows). The Orc6 subunit also functions in mitosis at kinetochores and during cytokinesis. Its detailed role in those processes is not known.
Replication Origins in Mammalian Cells
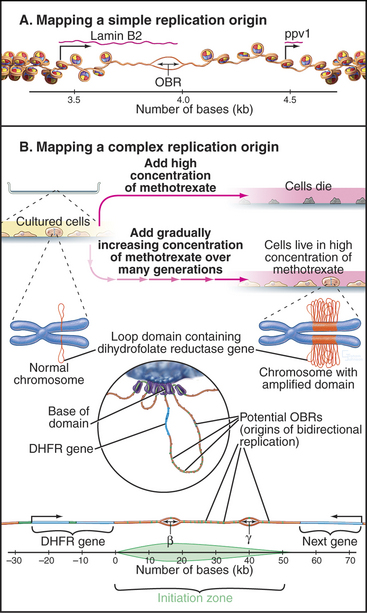
At present, two types of mammalian replication origins are known. The first is exemplified by the origin of replication adjacent to the lamin B2 gene (Fig. 42-6A). This origin “fires” within the first several minutes of the S phase, and a variety of methods have succeeded in mapping it to a stretch of less than 500 bp. Within this region, a single origin of bidirectional replication appears to be used. Thus, the lamin B2 origin of replication appears to be analogous to the well-characterized budding yeast origins.
The second is exemplified by the widely studied replication origin lying just downstream of the hamster gene for dihydrofolate reductase, an enzyme that is essential for biosynthesis of thymidine. This origin is accessible to experimental study because it is possible to select for cells with this chromosomal region amplified as hundreds or even thousands of copies (Fig. 42-6B). By looking for the first regions of the amplified DNA to replicate, the origin of replication was initially located within a region of about 55,000 bp. It now appears that DNA replication can initiate with low efficiency at roughly 20 sites distributed throughout this broad zone. Two of these sites are used with relatively higher efficiency, accounting for about 20% of all initiation in the region. These sites, termed Ori-β and Ori-γ (Fig. 42-6), each encompass about 0.5 to 2 kb of DNA.
Stay updated, free articles. Join our Telegram channel

Full access? Get Clinical Tree