17.1 INTRODUCTION TO RETROVIRUSES
The retroviruses are RNA viruses that copy their genomes into DNA during their replication. Until the discovery of these viruses it had been dogma that the transfer of genetic information always occurs in the direction of DNA to RNA, so finding that some viruses carry out “transcription backwards” (reverse transcription) caused something of a revolution. We now know that reverse transcription is carried out not only by these RNA viruses but also by some DNA viruses (see Chapter 19) and by uninfected cells.
The aim of the current chapter is to provide a general introduction to the retroviruses, which have been found in all classes of vertebrate animal, including fish, amphibians, birds, and mammals. The human immunodeficiency viruses (HIV-1 and HIV-2) are retroviruses and the next chapter is devoted entirely to these viruses. Many retroviruses can cause cancer in their hosts, and some aspects of this are discussed in Chapter 23.
17.2 RETROVIRUS VIRION
Retroviruses are unusual in that the virion contains two copies of the genome (Figure 17.1). The two RNA molecules are present as a dimer, formed by base pairing between complementary sequences. As well as the virus RNA, the virion also contains molecules of host cell RNA that were packaged during assembly. This host RNA includes a molecule of transfer RNA (tRNA) bound to each copy of the virus RNA through base pairing. The sequence in the virus RNA that binds a tRNA is known as the primer binding site (PBS). Each retrovirus binds a specific tRNA (Table 17.1).
Figure 17.1 Retrovirus virion and genome organization
| Genes | gag | group-specific antigen |
| pol | polymerase | |
| env | envelope | |
| Proteins | CA | capsid |
| IN | integrase | |
| MA | matrix | |
| NC | nucleocapsid | |
| PR | protease | |
| RH | ribonuclease H | |
| RT | reverse transcriptase | |
| SU | surface glycoprotein | |
| TM | transmembrane glycoprotein | |
| Sequences with roles in replication | PBS | primer binding site |
| PPT | polypurine tract | |
| R | repeat sequence | |
| U3 | unique sequence at 3′ end of genome | |
| U5 | unique sequence at 5′ end of genome | |
| ψ | Greek letter psi, from packaging signal |
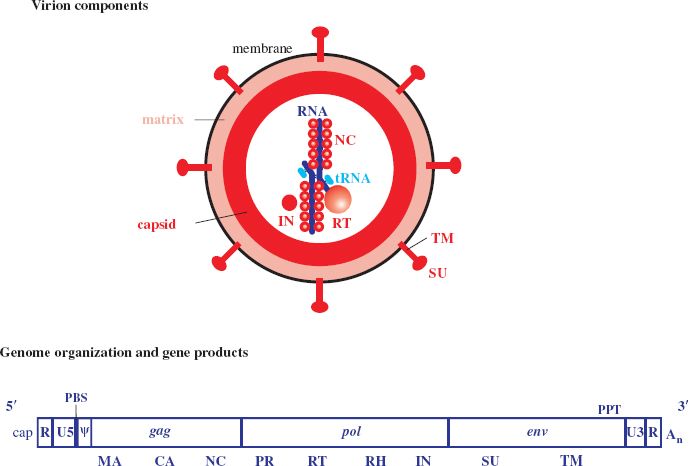
Table 17.1 Examples of tRNAs used by retroviruses as primers
| tRNA | Retrovirus |
| tRNApro | Human T-lymphotropic viruses 1 and 2 Murine leukemia virus |
| tRNAlys3 | HIV-1 and 2 Mouse mammary tumor virus |
A number of protein species are associated with the RNA. The most abundant protein is the nucleocapsid (NC) protein, which coats the RNA, while other proteins, present in much smaller amounts, have enzyme activities (Table 17.2).
Table 17.2 Enzyme activities present in the retrovirus virion
| RNA-dependent DNA polymerase (reverse transcriptase; RT) |
| DNA-dependent DNA polymerase |
| Ribonuclease H (RNase H) |
| Integrase |
| Protease |
Encasing the RNA and its associated proteins is the capsid, constructed from a lattice of capsid (CA) protein. The capsid has the shape of a sphere, a cone, or a cylinder, depending on the virus. A layer of matrix (MA) protein lies between the capsid and the envelope. Associated with the envelope are two protein species: a transmembrane (TM) protein bound to a heavily glycosylated surface (SU) protein. In most retroviruses the bonds between the TM and SU proteins are non-covalent. The TM–SU proteins are present in the envelope in trimers (groups of three).
The genes encoding the virus proteins are organized in three major regions of the genome (Figure 17.1):
- gag (group-specific antigen)—internal structural proteins;
- pol (pol ymerase)—enzymes;
- env (envelope)—envelope proteins.
17.3 RETROVIRUS REPLICATION
17.3.1 Attachment and entry
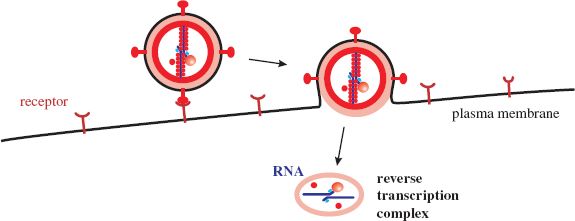
A site on the envelope SU protein binds to a cell receptor. This interaction causes a conformational change in the TM protein that allows a hydrophobic fusion sequence to fuse the virion membrane and a cell membrane (Section 5.2.4.b). Most retroviruses fuse their membrane with the plasma membrane of the cell (Figure 17.2), though some are endocytosed and fuse their membrane with an endosome membrane. The structure that is released into the cytoplasm loses some proteins and a reverse transcription complex is formed.
Figure 17.2 Retrovirus attachment and entry. Fusion of the virion membrane with the plasma membrane of the cell releases the virion contents, which undergo modification to form a reverse transcription complex.
17.3.2 Reverse transcription
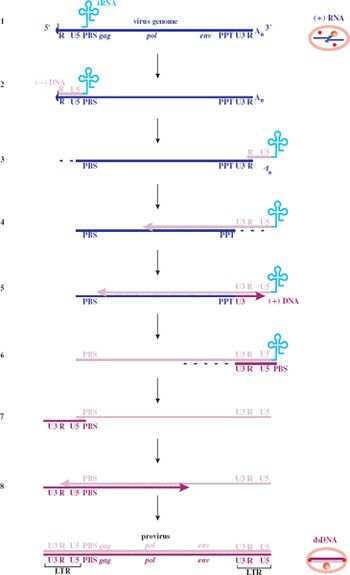
Reverse transcription takes place within the reverse transcription complex; some of the detail is indicated in Figure 17.3. Synthesis of both the (–) DNA and the (+) DNA begins at the 3′—OH of a primer RNA. The primer for synthesis of the (–) DNA is the tRNA bound to the genome, while the primer for synthesis of the (+) DNA is a polypurine tract (PPT) in the virus genome. The latter becomes accessible as a result of hydrolysis of the genome RNA from the 3′ end by the RNase H, which is an enzyme that specifically digests RNA in RNA–DNA duplexes.
Figure 17.3 Retroviral reverse transcription. LTR: long terminal repeat. PBS: primer binding site. PPT: polypurine tract (a sequence made up entirely, or almost entirely, of purine residues). R: repeat sequence. U3: unique sequence at 3′ end of genome. U5: unique sequence at 5′ end of genome.
1. A copy of the virus genome with a tRNA bound at the PBS.
2. The reverse transcriptase begins (–) DNA synthesis at the 3′ end of the tRNA.
3. The RNase H digests the RNA from the RNA–DNA duplex. The (–) DNA attaches at the 3′ end of either the same RNA strand or the second copy of the genome.
4. Elongation of the (–) DNA continues, while the RNase H degrades the template RNA from the 3′ end as far as the PPT.
5. Synthesis of (+) DNA begins.
6. The remaining RNA is degraded.
7. The (+) DNA detaches from the 5′ end of the (–) DNA template and attaches at the 3′ end.
8. Synthesis of both DNA strands is completed.
Stay updated, free articles. Join our Telegram channel

Full access? Get Clinical Tree


