INTRODUCTION
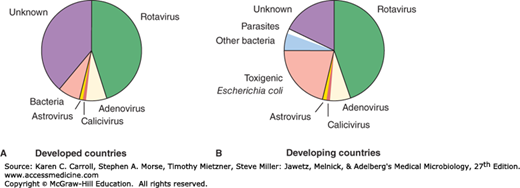
Reoviruses are medium-sized viruses with a double-stranded, segmented RNA genome. The family includes human rotaviruses, the most important cause of infantile gastroenteritis around the world (Figure 37-1). Acute gastroenteritis is a very common disease with significant public health impact. In developing countries, it is estimated to cause as many as 1.5 million deaths of preschool children annually, of which rotavirus is responsible for about 600,000 deaths. In the United States, acute gastroenteritis is second only to acute respiratory infections as a cause of disease in families.
Caliciviruses are small viruses with a single-stranded RNA genome. The family contains noroviruses, the major cause of nonbacterial epidemic gastroenteritis worldwide. Astroviruses also cause gastroenteritis.
REOVIRUSES AND ROTAVIRUSES
Important properties of reoviruses are summarized in Table 37-1.
| Virion: Icosahedral, 60–80 nm in diameter, double capsid shell |
| Composition: RNA (15%), protein (85%) |
| Genome: Double-stranded RNA, linear, segmented (10–12 segments); total genome size 16–27 kbp |
| Proteins: Nine structural proteins; core contains several enzymes |
| Envelope: None (transient pseudoenvelope is present during rotavirus particle morphogenesis) |
| Replication: Cytoplasm; virions not completely uncoated |
Outstanding characteristics: Genetic reassortment occurs readily Rotaviruses are the major cause of infantile diarrhea Reoviruses are good models for molecular studies of viral pathogenesis |
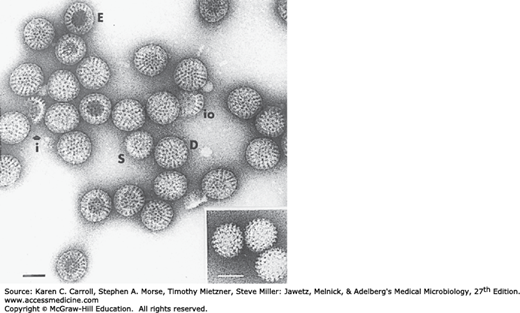
The virions measure 60–80 nm in diameter and possess two concentric capsid shells, each of which is icosahedral. (Rotaviruses have a triple-layered structure.) There is no envelope. Single-shelled virus particles that lack the outer capsid are 50–60 nm in diameter. The inner core of the particles is 33–40 nm in diameter (Figure 37-2). The double-shelled particle is the complete infectious form of the virus.
FIGURE 37-2
Electron micrograph of a negatively stained preparation of human rotavirus. D, double-shelled particles; E, empty capsids; i, fragment of inner shell; io, fragments of a combination of inner and outer shell; S, single-shelled particles. Inset: Single-shelled particles obtained by treatment of the viral preparation with sodium dodecyl sulfate. Bars, 50 nm. (Courtesy of J Esparza and F Gil.)
The reovirus genome consists of double-stranded RNA in 10–12 discrete segments with a total genome size of 16–27 kbp, depending on the genus. Whereas rotaviruses contain 11 genome segments, orthoreoviruses and orbiviruses each possess 10 segments and coltiviruses have 12 segments. The individual RNA segments vary in size from 680 (rotavirus) to 3900 bp (orthoreovirus). The virion core contains several enzymes needed for transcription and capping of viral RNAs.
Rotaviruses are stable to heat at 50°C, to a 3.0–9.0 range of pH, and to lipid solvents, such as ether and chloroform, but they are inactivated by 95% ethanol, phenol, and chlorine. Limited treatment with proteolytic enzymes increases infectivity.
The family Reoviridae is divided into 15 genera. Four of the genera are able to infect humans: Orthoreovirus, Rotavirus, Coltivirus, and Orbivirus. The genera are divided into two subfamilies: Spinareovirinae contains viruses with large spikes at the 12 vertices on the particle (eg, Orthoreovirus), whereas members of the Sedoreovirinae appear more smooth, lacking the large surface projections (eg, Rotavirus).
There are at least seven species or groups of rotaviruses (A–G) plus one recently proposed group (H), of which three species (A, B, C) infect humans. Strains of human and animal origin may fall in the same serotype. Other rotavirus groups and serotypes are found only in animals. Three different serotypes of reovirus are recognized, along with about 100 different orbivirus serotypes and two coltivirus serotypes.
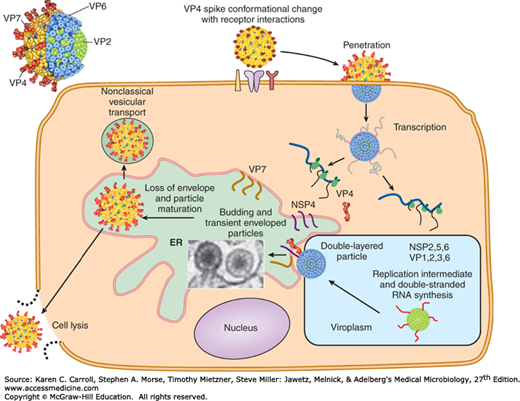
Viral particles attach to specific receptors on the cell surface (Figure 37-3). The cell attachment protein for reoviruses is the viral hemagglutinin (σ1 protein), a minor component of the outer capsid.
After attachment and penetration, uncoating of virus particles occurs in lysosomes in the cell cytoplasm. Only the outer shell of the virus is removed, and a core-associated RNA transcriptase is activated. This transcriptase transcribes mRNA molecules from the minus strand of each genome double-stranded RNA segment contained in the intact core. There are short terminal sequences at both ends of the RNA segments that are conserved among all isolates of a given subgroup. These conserved sequences may be recognition signals for the viral transcriptase. The functional mRNA molecules correspond in size to the genome segments. Most RNA segments encode a single protein, although a few (depending on the virus) encode two proteins. Reovirus cores contain all enzymes necessary for transcribing, capping, and extruding the mRNAs from the core, leaving the double-stranded RNA genome segments inside.
After being extruded from the core, the mRNAs are translated into primary gene products. Some of the full-length transcripts are encapsidated to form immature virus particles. A viral replicase is responsible for synthesizing negative-sense strands to form the double-stranded genome segments. This replication to form progeny double-stranded RNA occurs in partially completed core structures. The mechanisms that ensure assembly of the correct complement of genome segments into a developing viral core are unknown. However, genome reassortment occurs readily in cells coinfected with different viruses of the same subgroup, giving rise to virus particles containing RNA segments from the different parental strains. Viral polypeptides probably self-assemble to form the inner and outer capsid shells.
Reoviruses produce inclusion bodies in the cytoplasm in which virus particles are found. These viral factories are closely associated with tubular structures (microtubules and intermediate filaments). Rotavirus morphogenesis involves budding of single-shelled particles into the rough endoplasmic reticulum. The “pseudoenvelopes” so acquired are then removed and the outer capsids are added (Figure 37-3). This unusual pathway is used because the major outer capsid protein of rotaviruses is glycosylated.
Cell lysis results in release of progeny virions.
ROTAVIRUSES
Rotaviruses are a major cause of diarrheal illness in human infants and young animals, including calves and piglets. Infections in adult humans and animals are also common. Among the rotaviruses are the agents of human infantile diarrhea, Nebraska calf diarrhea, epizootic diarrhea of infant mice, and SA11 virus of monkeys.
Rotaviruses resemble reoviruses in terms of morphology and strategy of replication.
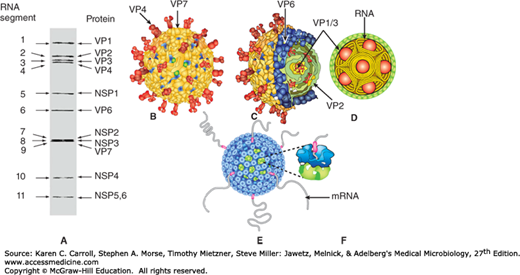
Rotaviruses have been classified into seven species (A–G) plus one tentative species (H) based on antigenic epitopes on the internal structural protein VP6. These can be detected by immunofluorescence, enzyme-linked immunosorbent assay (ELISA), and immune electron microscopy (IEM). Group A rotaviruses are the most frequent human pathogens. Outer capsid proteins VP4 and VP7 carry epitopes important in neutralizing activity, with VP7 glycoprotein being the predominant antigen. These type-specific antigens differentiate among rotaviruses and are demonstrable by neutralization tests. Five predominant serotype strains of rotavirus species A (G1-G4, G9) are responsible for the majority of human disease. Serotype distributions differ geographically. Multiple serotypes have been identified among human and animal rotaviruses. Some animal and human rotaviruses share serotype specificity. For example, monkey virus SA11 is antigenically very similar to human serotype 3. The gene-coding assignments responsible for the structural and antigenic specificities of rotavirus proteins are shown in Figure 37-4.
FIGURE 37-4
Rotavirus structure. A: Gel diagram showing the 11 segments of the genome. The structural (VP) and nonstructural (NSP) proteins encoded by these segments are indicated. B: Surface representation of the rotavirus structure from cryoelectron microscopic analysis. The two outer layer proteins are VP4, which forms the spikes, and VP7, which forms the capsid layer. C: Cut-away view showing the triple-layered organization of the virion, with the intermediate VP6 layer and the innermost VP2 layer indicated. The enzymes required for endogenous transcription (VP1) and capping (VP3) are attached as heterodimeric complexes to the inner surface of the VP2 layer. D: Proposed organization of the double-stranded RNA genome inside the VP2 layer along with transcription enzyme complexes (VP1/3) depicted as balls. E: Exit of transcripts from the channels at the fivefold vertices of actively transcribing double-layered particles. F: Close-up view of one of the exit channels. (Courtesy of BVV Prasad.)
Molecular epidemiologic studies have analyzed isolates based on differences in the migration of the 11 genome segments after electrophoresis of the RNA in polyacrylamide gels (Figure 37-5). These differences in electropherotypes can be used to differentiate species A viruses from other groups, but they cannot be used to determine serotypes.