Fig. 1
Phylogenetic tree of eubacteria (OTU  operational taxonomic unit) detected in infected cardiovascular devices. The phylogeny is based on the sequences of 16 S rRNA genes and calculated using the Maximum Likelihood algorithm
operational taxonomic unit) detected in infected cardiovascular devices. The phylogeny is based on the sequences of 16 S rRNA genes and calculated using the Maximum Likelihood algorithm
 operational taxonomic unit) detected in infected cardiovascular devices. The phylogeny is based on the sequences of 16 S rRNA genes and calculated using the Maximum Likelihood algorithm
operational taxonomic unit) detected in infected cardiovascular devices. The phylogeny is based on the sequences of 16 S rRNA genes and calculated using the Maximum Likelihood algorithm2.2 Biofilm Communities of Infected Implants
From 71 infected rhythm management devices DNA could be extracted from 95.8 % and the biofilm communities analyzed. As for the asymptomatic devices only biofilm communities were found and no mono-specific biofilms. Compared to the asymptomatic devices generally a higher biodiversity of the biofilm communities could be observed which were, contrary to the findings with the asymptomatic devices, dominated by Staphylococcus species. All devices with clinical signs of infection showed without exception the presence of Staphylococcus species. Due to the shortness of the 16rRNA gene sequences the underlying Staphylococcus species could not been identified. The sequence analyses identified Staphylococcus aureus or the closely related S. capitis or S. epidermidis as the main bacteria in all pathogenic biofilm communities [15]. This finding agreed perfectly with the microbiological analysis based on cultivation. Staphylococcus aureus, S. epidermidis and S. capitis are long known as the main pathogens in cardiovascular implant infections [16] but are these single- or a multi-pathogen infections and what is the role of the accompanying bacterial species?
To shed some light on this problem a phylogenetic analysis of the detected bacterial species were undertaken. Again a huge diversity of very different bacteria could be identified and is shown in a phylogenetic tree together with closest known species in Fig. 1. Remarkable is the large diversity of Pseudomonas species detected in the pathogenic biofilm communities but none of the identified Pseudomonas species has yet been connected with infections in humans. Again several rare bacterial species could be identified, among them Labrenzia, Leptotrichia [17], or Sphingobium species [18]. At this point it became interesting to compare the asymptomatic biofilm communities with the infectious ones. It turned out that in both groups Pseudomonas species were detected but any given Pseudomonas species occurred either in the asymptomatic or the infectious biofilm communities but never in both. A comparison of the occurrence of bacterial species in the biofilm communities led to the identification of species preferable present in asymptomatic biofilms while other species were indicators for infectious biofilm communities (Fig. 2). It is here interesting to note that two closely related bacterial species have been detected, here denoted Propionibacterium acnes 1 and 2. While P. acnes 1 was mainly found in asymptomatic biofilm communities, P. acnes 2 was a member of infectious biofilms. This observation underlines the importance of virulence factors which may play in some cases a more important role than the phylogeny of the bacteria.
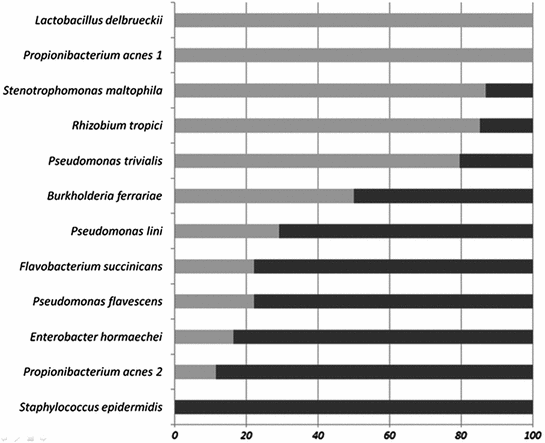

Fig. 2
The distribution of bacteria between asymptomatic (light grey) and infected devices (black bars)
2.3 Biofilms on Asymptomatic Implants in a Non-sterile Environment
The current understanding is that pacemakers or bone implants are placed into a sterile environment. But what happens to implants which are surrounded by bacteria? The skin, the gut or the mouth are always populated by a large number of bacteria, nevertheless, the mucosa and/or epithelial cells separate these microorganisms from the sterile tissues [19]. Their function as a barrier is of utmost importance for the entire organism. Implants for teeth breach this barrier and we became interested in their biofilms. As for the rhythm management devices we compared biofilm communities of asymptomatic and infected implants.
From ten patients samples of adherent supra- and subgingival periimplant biofilms were collected around titanium implants. Similar samples were also taken at remaining teeth for direct comparison of biofilms on implants and on teeth from the same patient. As for the pacemakers the microbial communities were analyzed using SSCP of 16 S rRNA gene amplicons. The diversity of Veillonella, Streptococcus, Parvimonas, Fusobacterium and Neisseria species detected was remarkable. The presence of various species of Streptococcus in the human mouth is well known and Streptococcus mitis and S. intermedius could be identified. However, no sequence belonging to the pathogen Streptococcus mutans was found confirming the asymptomatic nature of the biofilms. The genus Fusobacterium comprises strictly anaerobic bacteria and species of this genus are well known from the human mouth where they are important members of biofilm communities [20]. The species Fusobacterium naviforme and F. canifelinum were identified from the corresponding SSCP amplicons. Further well known members of asymptomatic dental biofilms, Neisseria mucosa and Parvimonas micra, were also detected in this study. As expected, a high diversity of bacteria was observed between biofilms of different patients [21]. Similar to the situation in the gut each individual seems to have in its dental biofilms its own microbial community. Applying multivariate analysis, however, revealed some bacteria species characteristic for subgingival or for supragingival biofilms. No discrimination, however, could be achieved between microbial species colonizing biofilms on implants or on residual teeth. This finding supports the view that biofilm communities on implants are not much different to the natural ones on teeth [22].
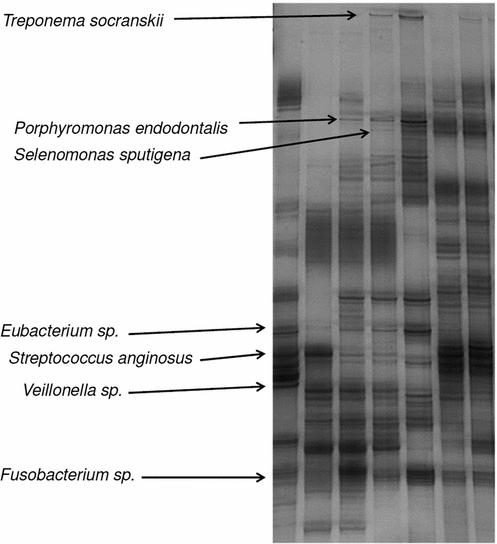

Fig. 3
Single strand conformation polymorphism (SSCP) gel of 16 S rRNA gene amplicons of biofilm communities from patients with peri-implantitis. Each lane stands for one biofilm community, each band is one distinct bacterial species. Bands migrating to the some height are identical species. Main bands have been cut out, their DNA sequenced and the species identified. Some of the identified bands are shown
2.4 Biofilms on Infected Dental Implants
Colonization of the implants by biofilms lead to peri-implantitis, an inflammation of peri-implant tissues, and is currently seen as one of the main causes for early implant failure. It is clinically characterized by inflammation of the mucosa and a subsequent destruction of the bone around the implant. To determine the differences in community composition between healthy and infected implants a similar analysis was performed for biofilm communities from patients with infected implants. But not only samples from the site of peri-implantitis but, where possible, also samples from asymptomatic implants and residual teeth from the same patients were taken and the biofilm communities analyzed.
As for the rhythm management devices a much higher diversity in bacterial species were detected in the pathogenic biofilm communities compared to their asymptomatic cousins (Fig. 3). Remarkable is here that species known for their anaerobic lifestyles dominated the biofilm communities indicating significant changes in the physiology of the biofilms. Socransky et al. assessed a large number of microbial communities from plaque samples and correlated them with clinical data. Using multivariate analyses they grouped the bacteria into five groups increasingly related to periodontitis [23]. These groups have also been applied in the assessment of peri-implantitis although its pathogenesis is somewhat different to the one of periodontitis. In our study no member of the bacterial group most closely related to periodontitis was observed but in some biofilm communities Streptococcus mutans, known to be the leading pathogen in the formation of dental cavities, was detected. In general was the diversity of Streptococcus and Fusobacterium species in peri-implantitis biofilms much higher than in asymptomatic ones. Comparing biofilm communities between different patients led again to the impression that each patient has his own set of dental biofilm communities. Surprising, however, was the observation that no large differences in the biofilm communities between infected and healthy implant of the same patient was found. The interpretation that pathogenic biofilm communities spread over all the teeth of an individual was supported by the finding that microbial communities from infected implants and healthy residual teeth were also similar. This leaves the question open why there is only infection on one implant and none on the others although the microbial communities are more or less the same. One interpretation could be that the barrier function of the gingival has been damaged and bacteria are reaching now areas where they cause inflammation and severe immune responses. A similar scenario has been proposed as the cause for inflammatory bowel disease [24].
Stay updated, free articles. Join our Telegram channel

Full access? Get Clinical Tree


