37
Protein Synthesis & the Genetic Code
OBJECTIVES
After studying this chapter, you should be able to:
![]() Understand that the genetic code is a three-letter nucleotide code, which is encoded in the linear array of the exon DNA (composed of triplets of A, G, C, and T) of protein coding genes, and that this three-letter code is translated into mRNA (composed of triplets of A, G, C, and U) to specify the linear order of amino acid addition during protein synthesis via the process of translation.
Understand that the genetic code is a three-letter nucleotide code, which is encoded in the linear array of the exon DNA (composed of triplets of A, G, C, and T) of protein coding genes, and that this three-letter code is translated into mRNA (composed of triplets of A, G, C, and U) to specify the linear order of amino acid addition during protein synthesis via the process of translation.
![]() Appreciate that the universal genetic code is degenerate, unambiguous, nonoverlapping, and punctuation free.
Appreciate that the universal genetic code is degenerate, unambiguous, nonoverlapping, and punctuation free.
![]() Explain that the genetic code is composed of 64 codons, 61 of which encode amino acids while 3 induce the termination of protein synthesis.
Explain that the genetic code is composed of 64 codons, 61 of which encode amino acids while 3 induce the termination of protein synthesis.
![]() Explain how the transfer RNAs (tRNAs) serve as the ultimate informational agents that decode the genetic code of mRNAs.
Explain how the transfer RNAs (tRNAs) serve as the ultimate informational agents that decode the genetic code of mRNAs.
![]() Understand the mechanism of the energy-intensive process of protein synthesis that occurs on RNA-protein complexes termed ribosomes.
Understand the mechanism of the energy-intensive process of protein synthesis that occurs on RNA-protein complexes termed ribosomes.
![]() Appreciate that protein synthesis, like DNA replication and transcription, is precisely controlled through the action of multiple accessory factors that are responsive to multiple extra- and intracellular regulatory signaling inputs.
Appreciate that protein synthesis, like DNA replication and transcription, is precisely controlled through the action of multiple accessory factors that are responsive to multiple extra- and intracellular regulatory signaling inputs.
BIOMEDICAL IMPORTANCE
The letters A, G, T, and C correspond to the nucleotides found in DNA. Within the protein-coding genes, these nucleotides are organized into three-letter code words called codons, and the collection of these codons makes up the genetic code. It was impossible to understand protein synthesis—or to explain mutations—before the genetic code was elucidated. The code provides a foundation for explaining the way in which protein defects may cause genetic disease and for the diagnosis and perhaps the treatment of these disorders. In addition, the pathophysiology of many viral infections is related to the ability of these infectious agents to disrupt host cell protein synthesis. Many antibacterial drugs are effective because they selectively disrupt protein synthesis in the invading bacterial cell but do not affect protein synthesis in eukaryotic cells.
GENETIC INFORMATION FLOWS FROM DNA TO RNA TO PROTEIN
The genetic information within the nucleotide sequence of DNA is transcribed in the nucleus into the specific nucleotide sequence of an RNA molecule. The sequence of nucleotides in the RNA transcript is complementary to the nucleotide sequence of the template strand of its gene in accordance with the base-pairing rules. Several different classes of RNA combine to direct the synthesis of proteins.
In prokaryotes there is a linear correspondence between the gene, the messenger RNA (mRNA) transcribed from the gene, and the polypeptide product. The situation is more complicated in higher eukaryotic cells, in which the primary transcript is much larger than the mature mRNA. The large mRNA precursors contain coding regions (exons) that will form the mature mRNA and long intervening sequences (introns) that separate the exons. The mRNA is processed within the nucleus, and the introns, which make up much more of this RNA than the exons, are removed. Exons are spliced together to form mature mRNA, which is transported to the cytoplasm, where it is translated into protein.
The cell must possess the machinery necessary to translate information accurately and efficiently from the nucleotide sequence of an mRNA into the sequence of amino acids of the corresponding specific protein. Clarification of our understanding of this process, which is termed translation, awaited deciphering of the genetic code. It was realized early that mRNA molecules themselves have no affinity for amino acids and, therefore, that the translation of the information in the mRNA nucleotide sequence into the amino acid sequence of a protein requires an intermediate adapter molecule. This adapter molecule must recognize a specific nucleotide sequence on the one hand as well as a specific amino acid on the other. With such an adapter molecule, the cell can direct a specific amino acid into the proper sequential position of a protein during its synthesis as dictated by the nucleotide sequence of the specific mRNA. In fact, the functional groups of the amino acids do not themselves actually come into contact with the mRNA template.
THE NUCLEOTIDE SEQUENCE OF AN mRNA MOLECULE CONTAINS A SERIES OF CODONS THAT SPECIFY THE AMINO ACID SEQUENCE OF THE ENCODED PROTEIN
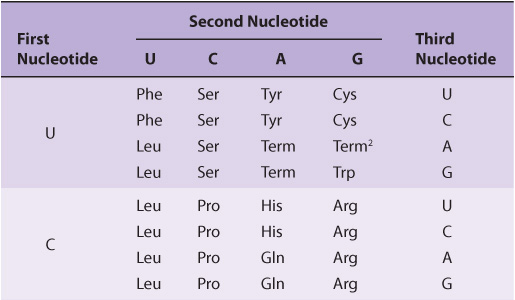
Twenty different amino acids are required for the synthesis of the cellular complement of proteins; thus, there must be at least 20 distinct codons that make up the genetic code. Since there are only four different nucleotides in mRNA, each codon must consist of more than a single purine or pyrimidine nucleotide. Codons consisting of two nucleotides each could provide for only 16 (42)-specific codons, whereas codons of three nucleotides could provide 64 (43)-specific codons.
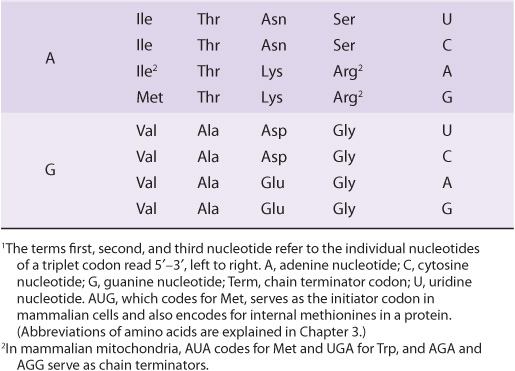
It is now known that each codon consists of a sequence of three nucleotides; that is, it is a triplet code (see Table 37-1). The initial deciphering of the genetic code depended heavily on in vitro synthesis of nucleotide polymers, particularly triplets in repeated sequence. These synthetic triplet ribonucleotides were used as mRNAs to program protein synthesis in the test tube, and allowed investigators to deduce the genetic code.
TABLE 37–1 The Genetic Code1 (Codon Assignments in Mammalian Messenger RNAs)
THE GENETIC CODE IS DEGENERATE, UNAMBIGUOUS, NONOVERLAPPING, WITHOUT PUNCTUATION, & UNIVERSAL
Three of the 64 possible codons do not code for specific amino acids; these have been termed nonsense codons. These nonsense codons are utilized in the cell as termination signals; they specify where the polymerization of amino acids into a protein molecule is to stop. The remaining 61 codons code for the 20 naturally occurring amino acids (Table 37-1). Thus, there is “degeneracy” in the genetic code—that is, multiple codons decode the same amino acid. Some amino acids are encoded by several codons; for example six different codons, UCU, UCC, UCA, UCG, AGU, and AGC all specify serine. Other amino acids, such as methionine and tryptophan, have a single codon. In general, the third nucleotide in a codon is less important than the first two in determining the specific amino acid to be incorporated, and this accounts for most of the degeneracy of the code. However, for any specific codon, only a single amino acid is indicated; with rare exceptions, the genetic code is unambiguous—that is, given a specific codon, only a single amino acid is indicated. The distinction between ambiguity and degeneracy is an important concept.
The unambiguous but degenerate code can be explained in molecular terms. The recognition of specific codons in the mRNA by the tRNA adapter molecules is dependent upon their anticodon region and specific base-pairing rules. Each tRNA molecule contains a specific sequence, complementary to a codon, which is termed its anticodon. For a given codon in the mRNA, only a single species of tRNA molecule possesses the proper anticodon. Since each tRNA molecule can be charged with only one specific amino acid, each codon therefore specifies only one amino acid. However, some tRNA molecules can utilize the anticodon to recognize more than one codon. With few exceptions, given a specific codon, only a specific amino acid will be incorporated—although, given a specific amino acid, more than one codon may be used.
As discussed below, the reading of the genetic code during the process of protein synthesis does not involve any overlap of codons. Thus, the genetic code is nonoverlapping. Furthermore, once the reading is commenced at a specific codon, there is no punctuation between codons, and the message is read in a continuing sequence of nucleotide triplets until a translation stop codon is reached.
Until recently, the genetic code was thought to be universal. It has now been shown that the set of tRNA molecules in mitochondria (which contain their own separate and distinct set of translation machinery) from lower and higher eukaryotes, including humans, reads four codons differently from the tRNA molecules in the cytoplasm of even the same cells. As noted in Table 37-1, the codon AUA is read as Met, and UGA codes for Trp in mammalian mitochondria. In addition, in mitochondria, the codons AGA and AGG are read as stop or chain terminator codons rather than as Arg. As a result of these organelle-specific changes in genetic code, mitochondria require only 22 tRNA molecules to read their genetic code, whereas the cytoplasmic translation system possesses a full complement of 31 tRNA species. These exceptions noted, the genetic code is universal. The frequency of use of each amino acid codon varies considerably between species and among different tissues within a species. The specific tRNA levels generally mirror these codon usage biases. Thus, a particular abundantly used codon is decoded by a similarly abundant-specific tRNA which recognizes that particular codon. Tables of codon usage are becoming more accurate as more genes and genomes are sequenced; such information can prove vital for large-scale production of proteins for therapeutic purposes (ie, insulin, erythropoietin). Such proteins are often produced in nonhuman cells using recombinant DNA technology (Chapter 39). The main features of the genetic code are listed in Table 37-2.
TABLE 37–2 Features of the Genetic Code
AT LEAST ONE SPECIES OF TRANSFER RNA (tRNA) EXISTS FOR EACH OF THE 20 AMINO ACIDS
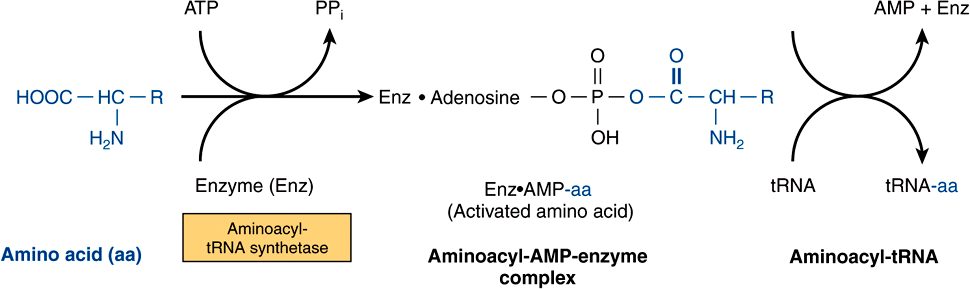
tRNA molecules have extraordinarily similar functions and three-dimensional structures. The adapter function of the tRNA molecules requires the charging of each specific tRNA with its specific amino acid. Since there is no affinity of nucleic acids for specific functional groups of amino acids, this recognition must be carried out by a protein molecule capable of recognizing both a specific tRNA molecule and a specific amino acid. At least 20-specific enzymes are required for these specific recognition functions and for the proper attachment of the 20 amino acids to specific tRNA molecules. The energy requiring process of recognition and attachment (charging) proceeds in two steps and is catalyzed by one enzyme for each of the 20 amino acids. These enzymes are termed aminoacyl-tRNA synthetases. They form an activated intermediate of aminoacyl-AMP-enzyme complex (Figure 37–1). The specific aminoacyl-AMP-enzyme complex then recognizes a specific tRNA to which it attaches the aminoacyl moiety at the 3′-hydroxyl adenosyl terminal. The charging reactions have an error rate of less than 10-4 and so are quite accurate. The amino acid remains attached to its specific tRNA in an ester linkage until it is polymerized at a specific position in the fabrication of a polypeptide precursor of a protein molecule.
FIGURE 37–1 Formation of aminoacyl-tRNA. A two-step reaction, involving the enzyme amino-acyl-tRNA synthetase, results in the formation of aminoacyl-tRNA. The first reaction involves the formation of an AMP-amino acid-enzyme complex. This activated amino acid is next transferred to the corresponding tRNA molecule. The AMP and enzyme are released, and the latter can be reutilized. The charging reactions have an error rate (ie, esterifying the incorrect amino acid on tRNAx) of less than 10-4.
The regions of the tRNA molecule referred to in Chapter 34 (and illustrated in Figure 34–11) now become important. The ribothymidine pseudouridine cytidine (TψC) arm is involved in binding of the aminoacyl-tRNA to the ribosomal surface at the site of protein synthesis. The D arm is one of the sites important for the proper recognition of a given tRNA species by its proper aminoacyl-tRNA synthetase. The acceptor arm, located at the 3′-hydroxyl adenosyl terminal, is the site of attachment of the specific amino acid.
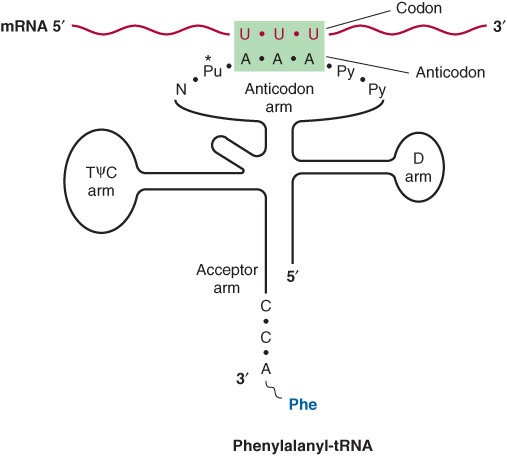
The anticodon region consists of seven nucleotides, and it recognizes the three-letter codon in mRNA (Figure 37–2). The sequence read from the 3′-5’ direction in that anticodon loop consists of a variable base-modified purine-XYZ-pyrimidine-pyrimidine-5’. Note that this direction of reading the anticodon is 3′-5’, whereas the genetic code in Table 37-1 is read 5′-3’, since the codon and the anticodon loop of the mRNA and tRNA molecules, respectively, are antiparallel in their complementarity just like all other intermolecular interactions between nucleic acid strands.
FIGURE 37–2 Recognition of the codon by the anticodon. One of the codons for phenylalanine is UUU. tRNA charged with phenylalanine (Phe) has the complementary sequence AAA; hence, it forms a base-pair complex with the codon. The anticodon region typically consists of a sequence of seven nucleotides: variable (N), modified purine (Pu*), X, Y, Z (here, A A A), and two pyrimidines (Py) in the 3′ -5’ direction.
The degeneracy of the genetic code resides mostly in the last nucleotide of the codon triplet, suggesting that the base pairing between this last nucleotide and the corresponding nucleotide of the anticodon is not strictly by the Watson-Crick rule. This is called wobble; the pairing of the codon and anti-codon can “wobble” at this specific nucleotide-to-nucleotide pairing site. For example, the two codons for arginine, AGA and AGG, can bind to the same anticodon having a uracil at its 5′ end (UCU). Similarly, three codons for glycine—GGU, GGC, and GGA—can form a base pair from one anticodon, 3′ CCI 5′ (ie, I can base pair with U, C and A). I is a purine inosine nucleotide generated by deamination of adenine (see Figure 33–2 for structure), another of the peculiar bases often appearing in tRNA molecules.
MUTATIONS RESULT WHEN CHANGES OCCUR IN THE NUCLEOTIDE SEQUENCE
Although the initial change may not occur in the template strand of the double-stranded DNA molecule for that gene, after replication, daughter DNA molecules with mutations in the template strand will segregate and appear in the population of organisms.
Some Mutations Occur by Base Substitution
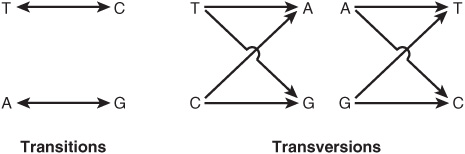
Single-base changes (point mutations) may be transitions or transversions. In the former, a given pyrimidine is changed to the other pyrimidine or a given purine is changed to the other purine. Transversions are changes from a purine to either of the two pyrimidines or the change of a pyrimidine into either of the two purines, as shown in Figure 37–3.
FIGURE 37–3 Diagrammatic representation of transition mutations and transversion mutations.
If the nucleotide sequence of the gene containing the mutation is transcribed into an RNA molecule, then the RNA molecule will of course possess the base change at the corresponding location.
Single-base changes in the mRNA molecules may have one of several effects when translated into protein:
1. There may be no detectable effect because of the degeneracy of the code; such mutations are often referred to as silent mutations. This would be more likely if the changed base in the mRNA molecule were to be at the third nucleotide of a codon. Because of wobble, the translation of a codon is least sensitive to a change at the third position.
2. A missense effect will occur when a different amino acid is incorporated at the corresponding site in the protein molecule. This mistaken amino acid—or missense, depending upon its location in the specific protein—might be acceptable, partially acceptable, or unacceptable to the function of that protein molecule. From a careful examination of the genetic code, one can conclude that most single-base changes would result in the replacement of one amino acid by another with rather similar functional groups. This is an effective mechanism to avoid drastic change in the physical properties of a protein molecule. If an acceptable missense effect occurs, the resulting protein molecule may not be distinguishable from the normal one. A partially acceptable missense will result in a protein molecule with partial but abnormal function. If an unacceptable missense effect occurs, then the protein molecule will not be capable of functioning normally.
3. A nonsense codon may appear that would then result in the premature termination of amino acid incorporation into a peptide chain and the production of only a fragment of the intended protein molecule. The probability is high that a prematurely terminated protein molecule or peptide fragment will not function in its assigned role. Examples of the different types of mutations, and their effects on the coding potential of mRNA are shown in Figures 37-4 and 37-5.
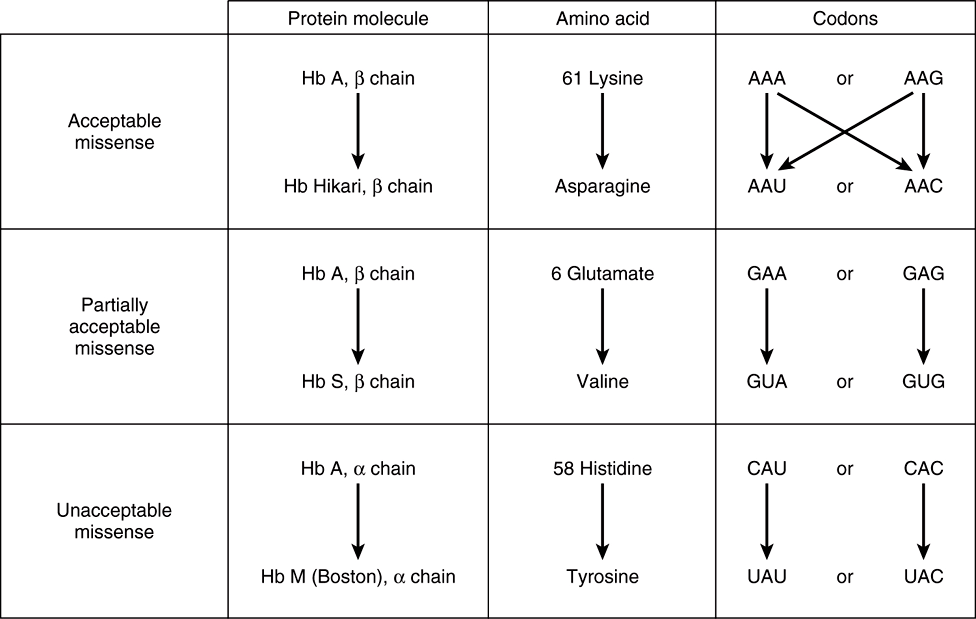
FIGURE 37–4 Examples of three types of missense mutations resulting in abnormal hemoglobin chains. The amino acid alterations and possible alterations in the respective codons are indicated. The hemoglobin Hikari β-chain mutation has apparently normal physiologic properties but is electrophoretically altered. Hemoglobin S has a β-chain mutation and partial function; hemoglobin S binds oxygen but precipitates when deoxygenated; this causes red blood cells to sickle, and represents the cellular and molecular basis of sickle cell disease (see Figure 6–12). Hemoglobin M Boston, an α-chain mutation, permits the oxidation of the heme ferrous iron to the ferric state and so will not bind oxygen at all.
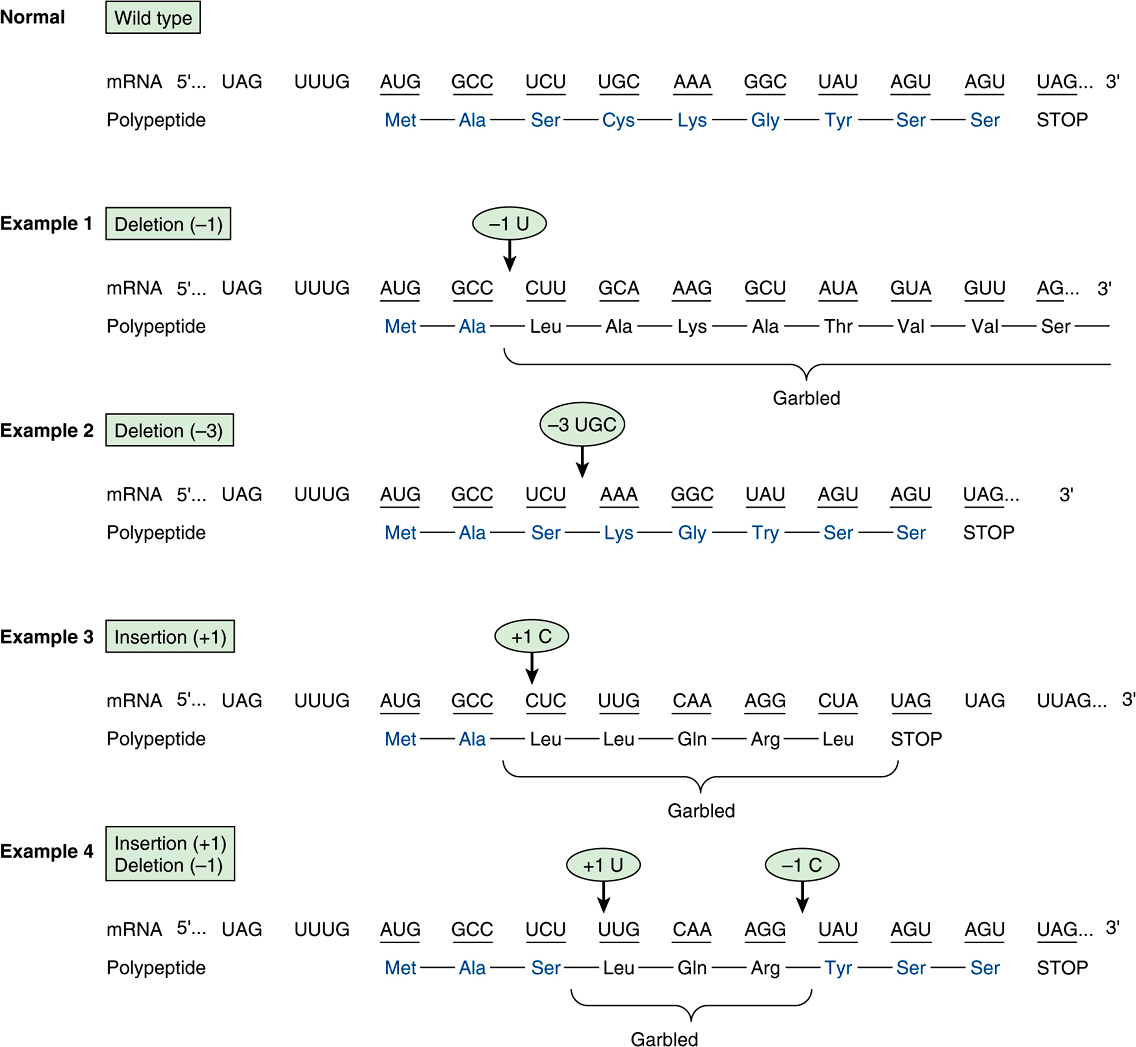
FIGURE 37–5 Examples of the effects of deletions and insertions in a gene on the sequence of the mRNA transcript and of the polypeptide chain translated therefrom. The arrows indicate the sites of deletions or insertions, and the numbers in the ovals indicate the number of nucleotide residues deleted or inserted. Colored type indicates amino acids in correct order.
Stay updated, free articles. Join our Telegram channel

Full access? Get Clinical Tree