CHAPTER 17 Protein Synthesis and Folding*
The nuclear genome contains information specifying many thousands of proteins. Whatever their final destination—nucleus, cytoplasm, membrane-bound organelles, or extracellular space—these proteins are synthesized in the cytoplasm. The few proteins encoded by genes in mitochondria and chloroplasts are synthesized in those organelles. The biochemical synthesis of proteins is called translation, as the process translates sequences of nucleotides in a messenger RNA (mRNA) into the sequence of amino acids in a polypeptide chain. Translation of mRNA requires the concerted actions of small transfer RNAs (tRNAs) linked to amino acids, ribosomes (complexes of RNA and protein), and many soluble proteins. GTP binding and hydrolysis regulate several proteins that orchestrate the interactions of these components. Ultimately, RNA bases in the ribosome catalyze the formation of peptide bonds. Some newly synthesized polypeptides fold spontaneously into their native structure in the cellular environment, but many require assistance from proteins called chaperones. It has been proposed that the bulk of the evolution of the translation apparatus occurred after the basic mechanisms were established, to provide greater precision. This perspective seems to explain the extraordinary complexity of the process.
Protein Synthetic Machinery
Messenger RNA
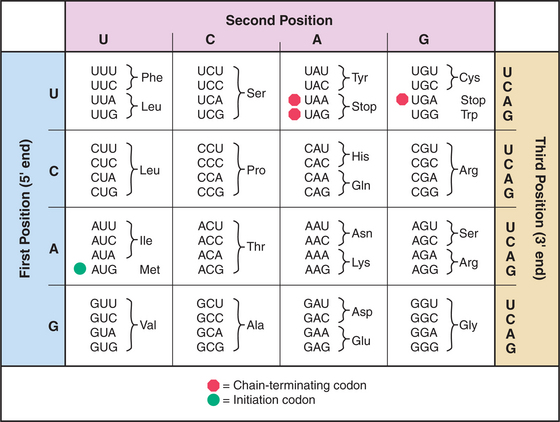
mRNAs have three parts: Nucleotides at the 5′ end provide binding sites for proteins that initiate polypeptide synthesis; nucleotides in the middle specify the sequence of amino acids in the polypeptide; and nucleotides at the 3′ end regulate the stability of the mRNA (see Figs. 15-1 and 16-1). Within the protein-coding region, successive triplets of three nucleotides, called codons, specify the sequence of amino acids. The genetic code relating nucleotide triplets to amino acids is, with a few minor exceptions, universal. One to six different triplet codons encode each amino acid (Fig. 17-1). An initiation codon (AUG) specifies methionine, which begins all polypeptide chains. In addition, any one of three termination codons (UAA, UGA, UAG) stops peptide synthesis.
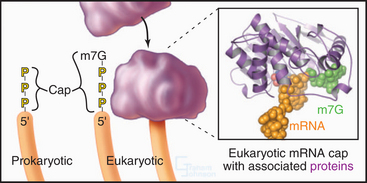
Eukaryotic and bacterial mRNAs differ in three ways. First, eukaryotic mRNAs encode one protein, and bacterial mRNAs generally encode more than one protein. Second, most eukaryotic (and eukaryotic viral) mRNAs are capped by an inverted 7-methylguanosine residue joined onto the 5′ end of the mRNA by a 5′-triphosphate-5′ linkage (Fig. 17-2). This 5′ cap is stable throughout the life of the mRNA and protects the 5′ end against attack by nucleases. Third, most eukaryotic mRNAs have a tail of 50 to 200 adenine residues added posttranscriptionally to the 3′ end (see Fig. 16-3). The poly(A) tail may protect the mRNA from degradation in the cytoplasm and increase reinitiation of transcription. Bacterial mRNAs lack 5′ caps or 3′ poly(A) tails. Most eukaryotic mRNAs require processing to remove introns (see Fig. 16-4). Many single-stranded mRNAs have some secondary structure (see Fig. 3-19) stabilized by hydrogen bonding of complementary bases. This secondary structure must be disrupted during translation to allow reading of each codon.
Transfer RNA
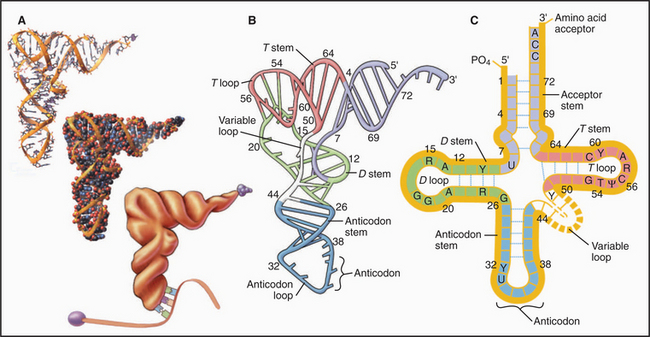
tRNAs are adapters that deliver amino acids to the translation machinery by matching mRNA codons with their corresponding amino acids as they are incorporated into a growing polypeptide (Fig. 17-3). One to four different tRNAs are specific for each amino acid, generally reflecting their abundance in proteins. Specialized tRNAs carrying methionine (formylmethionine in Bacteria) initiate protein synthesis. Transfer RNAs consist of about 76 nucleotides that base-pair to form four stems and three intervening loops. These elements of secondary structure fold to form an L-shaped molecule stabilized by base pairing. A “decoding” triplet (the anticodon) is at one end of the L (the anticodon arm), and the amino acid acceptor site is at the other end of the L (the acceptor arm).
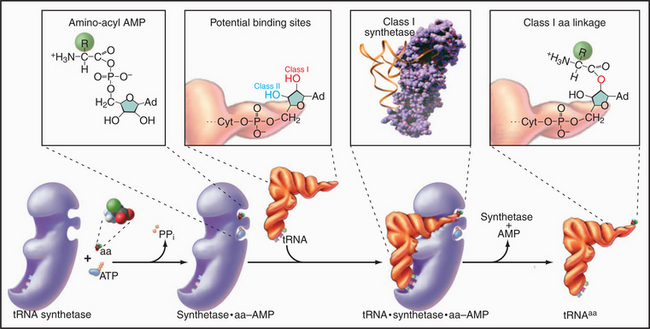
Enzymes called aminoacyl-tRNA (aa-tRNA) synthetases catalyze a two-step reaction that couples an amino acid covalently to its cognate tRNA but not to any other tRNA (Fig. 17-4). In the first step, adenosine triphosphate (ATP) and the amino acid react to form a high-energy aminoacyl adenosine monophosphate (AMP) intermediate with release of pyrophosphate. The second step transfers the amino acid to the 3′ adenine of tRNA, forming an aa-tRNA. This reaction is appropriately called charging, since the high-energy bond between the amino acid and the tRNA activates the amino acid in preparation for forming a peptide bond with an amino group in the growing polypeptide chain. Each of the 20 aa-tRNA synthetases couples a particular amino acid to all of its corresponding tRNAs.
The fidelity of protein synthesis depends on near-perfect coupling of amino acids to the appropriate tRNAs. Synthetases make this selection by interacting with as many as three areas of their cognate tRNAs: anticodon, 3′ acceptor stem, and the surface between these sites (Fig. 17-4). To distinguish between appropriate and inappropriate amino acids, synthetases use proofreading steps, which remove incorrectly paired amino acids from tRNAs.
Ribosomes
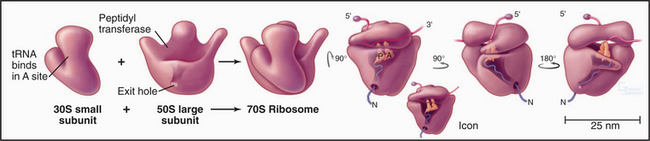
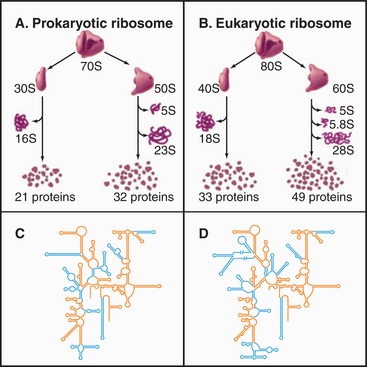
Ribosomes are giant macromolecular machines that bring together an mRNA and aa-tRNAs to synthesize a polypeptide. Base pairing between mRNA codons and tRNA anticodons directs the synthesis of a polypeptide in the order specified by the mRNA codons. Ribosomes consist of a small subunit and a large subunit that bind together during translation of an mRNA (Fig. 17-5). Each subunit consists of one or more ribosomal RNA (rRNA) molecules and many distinct proteins (Fig. 17-6). The sizes of these subunits and rRNAs are traditionally given in units of S, the sedimentation coefficient measured in an ultracentrifuge.
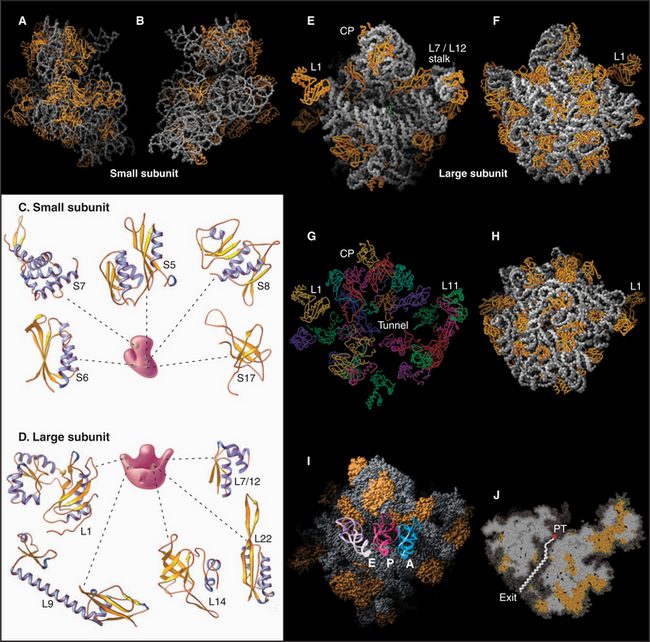
Ribosomal RNAs constitute the structural core of each ribosomal subunit (Fig. 17-7). The 16S rRNA of the small subunit consists of 1500 bases, most of which are folded into base-paired helices. The large subunit contains two RNAs: 23 S rRNA consisting of 2900 bases and 5S rRNA of 121 bases. The rRNAs fold into many based-paired helices, as predicted by phylogenetic analysis of sequences (Fig. 17-6). These helices and their intervening loops pack into a compact structure, as is seen in both surface views and cross sections. Although eukaryotic rRNAs differ in size and sequence from prokaryote rRNAs, their predicted secondary structures are similar, and they are expected to fold in similar ways. Many features of rRNAs have been conserved during evolution, including the surfaces where subunits and elements of RNA structure interact; sites that are required for binding tRNA, mRNA, and protein cofactors; and the residues involved with peptide bond formation.
Most ribosomal proteins associate with the surface of the rRNA core, although several extend peptide strands into the core (Fig. 17-7). Ribosomal proteins are generally small (10 to 30 kD) and basic, but each has a unique structure. With one exception, ribosomes have just one copy of each protein.
Outline of Protein Synthesis
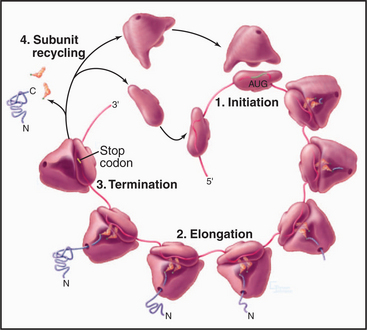
Organisms in all three domains of life use many homologous components and similar reactions for protein synthesis, but many of the details differ as is expected after 3 billion years of evolutionary divergence. In all three domains, protein synthesis takes place in four steps: initiation, elongation, termination, and subunit recycling (Fig. 17-8). Guanosine triphosphatase (GTPase) proteins regulate the progress and fidelity of many of the steps (see Fig. 4-6 for details on GTPase cycles). Initiation, elongation, and termination all depend on directed movement of molecular machinery along an mRNA and precise recognition between amino acids, tRNAs, adapter proteins, and the gene sequence encoded in the mRNA.
Stay updated, free articles. Join our Telegram channel

Full access? Get Clinical Tree