Principles of Cytogenetics*
Linda A. Cannizzaro
The events governing the developmental evolution of cells as they progress from the fertilized ovum to mature tissues are not fully understood as yet. It is known, however, that this process involves extensive proliferation and differentiation of embryonal stem cells and their selective destruction by programmed cell death or apoptosis (see Chap. 6). These processes are governed by messages inscribed in the nuclear deoxyribose nucleic acid (DNA) (see Chap. 3). The key feature in cell proliferation is cell division.
There are two forms of cell division, one occurring during the formation of gametes (e.g., the spermatozoa and ova), known as meiosis, and the other affecting all other cells (somatic cells) known as mitosis. The purpose of meiosis is to reduce the number of chromosomes by one half (in humans from 46 to 23) in the gametes, so that the union of a spermatozoon and an ovum (fertilization of the ovum) will result in an organism that carries the full complement of chromosomes (in humans, 46) in its somatic cells. The purpose of mitosis is the reproduction of somatic cells, each carrying the full complement of chromosomes. Both forms of cell division are discussed in this chapter.
The events encompassing the life of a cell from its birth until the end of the mitotic division are known as the cell cycle, during which the genomic identity of the cell, vested in the DNA, must be preserved. Molecular genetic technology has considerably advanced our knowledge of the processes involved in the progression of the cell cycle. The normal cell cycle has developed complex mechanisms for the detection and repair of damaged DNA. Upsetting the intricate balance of these cellular processes has dramatic and usually tragic consequences. Dysregulation of meiosis oftentimes is manifested as a genetic disorder, while dysregulation of mitosis may result in a malignant disorder.
Since the demonstration of the specificity of chromosomal changes in many disease states and their utilization in diagnosis, the cytogenetic aspects of human diseases have become of direct concern to the practicing physician. This chapter summarizes the salient features of cell division, as well as some of the inherited and malignant conditions that directly result from faulty or anomalous events during meiosis and mitosis. Recent introduction of several powerful molecular cytogenetic methods has facilitated the identification of chromosomal alterations previously irresolvable by high-resolution cytogenetic analysis. These technologies, including the recent mapping of the human genome (Caron et al, 2001; International Human Genome Sequencing Consortium, 2001; Venter et al, 2001; Peltonen and McKusick, 2001) have enormously impacted our knowledge of human genetic disease and the contributions made by these innovations will be made evident in the forthcoming narrative.
THE CELL CYCLE
The cell cycle is composed of several phases, which have, for their purpose, the preservation of the genomic heritage of the cell to be transmitted to the two daughter cells.
The phases of the cell cycle are as follows:
G0 (resting phase)
G1 (gap1)
S (synthesis)
G2 (gap2)
M (mitosis)
The events in the phases of cell cycle are described below.
Events Preparatory to Cell Division
Genetic information in the form of DNA is stored within the interphase nucleus in thread-like, tangled structures called chromatin. During the process of cell division, the DNA condenses and divides into several distinct pairs of linear segments or chromosomes. Each time the cell divides, the hereditary information carried in the chromosomes is passed on to the two newly formed cells. The DNA in the nucleus contains the instructions for regulating the amount and types of proteins made by the cell. These instructions are copied, or transcribed, into messenger RNA (mRNA), which is transported from the nucleus to the ribosomes located in the cytoplasm, where proteins are assembled (see Chap. 3).
Most somatic cells spend the greater part of their lives in G0, or the resting phase of the cell cycle, because such cell populations are not actively dividing.
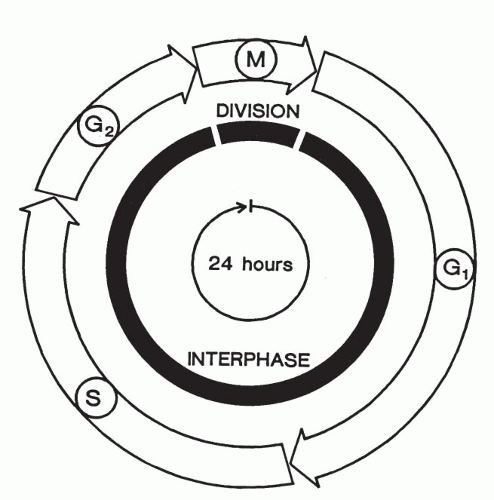
Before a cell can divide, it must double its mass and duplicate all of its contents. This ensures the ability of the daughter cells to begin their own cycle of growth followed by division. Most of the work involved in preparing for division goes on invisibly during the growth phase of the cell cycle, known as the interphase, which comprises the G1, S, and G2 phases of the cell cycle (Fig. 4-1). The interphase nucleus is the seat of crucial biochemical activities including the synthesis of proteins and the duplication of its chromosomal DNA in preparation for subsequent cell division.
Cell Division
The process of cell division (see Fig. 4-1) can be readily visualized in the microscope and consists of two sequential
events: nuclear division (mitosis) followed by cytoplasmic division (cytokinesis). The cell-division phase is designated as the M phase (M = mitosis). The period between the end of the M phase and the start of DNA synthesis is the G1 phase (G = gap). In G1, RNAs and proteins, including the essential components needed for DNA replication, are synthesized without replication of DNA. Once all the ingredients are synthesized in G1, DNA replication takes place in the ensuing synthesis phase (S-phase) of the cell cycle.
events: nuclear division (mitosis) followed by cytoplasmic division (cytokinesis). The cell-division phase is designated as the M phase (M = mitosis). The period between the end of the M phase and the start of DNA synthesis is the G1 phase (G = gap). In G1, RNAs and proteins, including the essential components needed for DNA replication, are synthesized without replication of DNA. Once all the ingredients are synthesized in G1, DNA replication takes place in the ensuing synthesis phase (S-phase) of the cell cycle.
The period between the completion of DNA synthesis and the M phase is known as the G2 phase, in which additional cellular components are synthesized in preparation for the cell’s entry into mitosis. The interphase thus consists of successive G1, S, and G2 phases that normally constitute 90% or more of the total cell cycle time (see Fig. 4-1).
However, following the completion of mitotic division, most normal somatic cells leave the division cycle and enter a postmitotic resting phase (G0), rather than the new G1 phase. The unknown trigger mechanism for cell division is activated during the G0 phase; as a result, the cell enters G1 phase and is committed to divide (Brachet, 1985; Levitan, 1987; Therman, 1993; Nicklas, 1997; Hixon and Gualberto, 2000). In fact, experiments have shown that the point of no return, known as the restriction point (R point), occurs late in G1. After cells have passed this point, they will complete the rest of the cycle at their normal rate, regardless of external conditions. The time spent by cells in G2 and S phases is relatively constant (Brachet, 1985; Gardner, 2000). One interesting exception is the epidermis of the skin, in which some cells remain in the G2 phase and thus are able to undergo rapid division in wound healing.
Studies of the cell cycle in yeast have shown that the cell proceeds from one phase of the cell cycle to the next by passing through a series of molecular checkpoints (Li and Murray, 1983). These checkpoints determine whether the cell is ready to enter into the next phase of the cell cycle. These biochemical checkpoints involve the synthesis of new proteins and degradation of already existing proteins. Both the S phase and the M phase are activated by related protein kinases, which function at specific stages of the cell cycle. Each kinase consists of at least two subunits, one of which is cyclin, so named because of its role in the cell cycle. There are several cyclins involved in regulating entry into different parts of the cell cycle, and they are degraded after serving their purpose or as the cell progresses in the cycle and through mitosis (Rudner and Murray, 1996; Amon, 1999; Cerrutti et al, 2000; Gardner, 2000).
The cells of the human body divide at very different rates. Some cells, such as mature neurons, heart and skeletal muscle, and mature red blood cells, do not divide at all or perhaps only under most exceptional circumstances. Other cells, such as the epithelial cells that line the inside and outside surfaces of the body (e.g., the intestine, lung, and skin), divide continuously and relatively rapidly throughout the life of the individual. The behavior of most cells falls somewhere between these two extremes. Most somatic cells rarely divide, and the duration of their cell cycle may be 100 days or more.
The average time for the mitotic cycle in most cell types is about 16 hours in human and other mammalian cells, distributed as follows: S phase, approximately 6 to 8 hours; G1 phase, 6 to 12 hours; G2 phase, 4 hours; and M phase, 1 to 2 hours (see Fig. 4-1). The M, and especially G1, phases may show considerable variation in duration. Most of the available evidence suggests that these periods are longer in cancer cells than in benign cells, or at least in benign cell populations that normally have a rapid turnover. Many tissues require more than 16 hours to complete the mitosis (Miles, 1979).
Even though it takes a minimum of 7 to 8 hours for a cell to duplicate its entire chromosomal DNA, individual chromosomes or segments of chromosomes are replicated asynchronously, some of them sooner and faster than others. Thus, some chromosomes, or their segments, will have completed DNA synthesis before others begin. This asynchrony does not follow a simple pattern. The synthesis does not necessarily begin at one point and spread uniformly along the chromosome, but may start at several places on a single chromosome, while others wait their turn for DNA replication. A reproducible phenomenon is the late replication of one of the two X chromosomes in normal female cells or in cells with more than one X chromosome. Apparently, this X chromosome finishes its DNA replication later than any other chromosome in the cell. The number of late-replicating X chromosomes is usually one less than the total number of X chromosomes in the cell (Moore, 1966; Sandberg, 1983a, 1983b).
The chromosomes are not visible under the light microscope except during the M phase of the cell cycle. The physical condition of the chromosomes during interphase (e.g., G1, S, and G2) is not known, but their invisibility is probably caused, at least in part, to their enormous elongation. The older notion that the chromosomes lose their linear structure and become dissolved in the nucleoplasm is unlikely, and it introduces unnecessary complexities into the analysis of nuclear and chromosomal dynamics (van Holde, 1989; Miles, 1964, 1979).
Recent studies of chromosomes, utilizing fluorescent probes for chromosomal “painting,” suggest that the chromosomes retain their distinct identity during the interphase and that their position in the nucleus may be relatively constant throughout the life of the cell (Nagele et al, 1995; Koss, 1998).
CHROMOSOME STRUCTURE
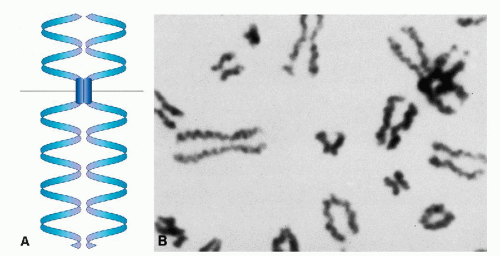
Soon after a chromosome becomes visible in the early part (prophase) of mitotic division, it is already doubled into a pair of identical chromatids (Fig. 4-2A). This pair remains joined together at one point, the centromere (also called the primary constriction). The centromere divides the chromosome into a short (from French, p = petit) and a long (q, the next letter after p) arm
region. The centromere connects the chromosome to the spindle fibers during mitotic division. Associated with the centromere are proteinaceous structures, known as kinetechores, to which the microtubules of the spindle mechanism are attached (see below). Normal chromosome ends are capped by telomeres. These short repeat DNA sequences are essential for maintaining the structural integrity of the chromosome by preventing the ends from fusing with other chromosomes. If the telomere sequences are lost or broken off, an end-to-end fusion of two chromosomes can occur.
region. The centromere connects the chromosome to the spindle fibers during mitotic division. Associated with the centromere are proteinaceous structures, known as kinetechores, to which the microtubules of the spindle mechanism are attached (see below). Normal chromosome ends are capped by telomeres. These short repeat DNA sequences are essential for maintaining the structural integrity of the chromosome by preventing the ends from fusing with other chromosomes. If the telomere sequences are lost or broken off, an end-to-end fusion of two chromosomes can occur.
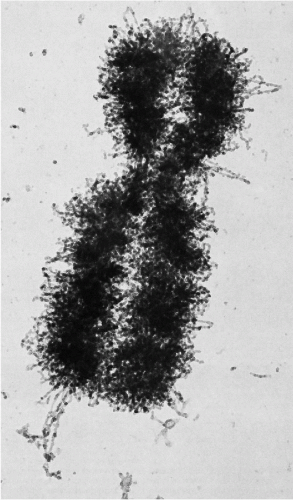
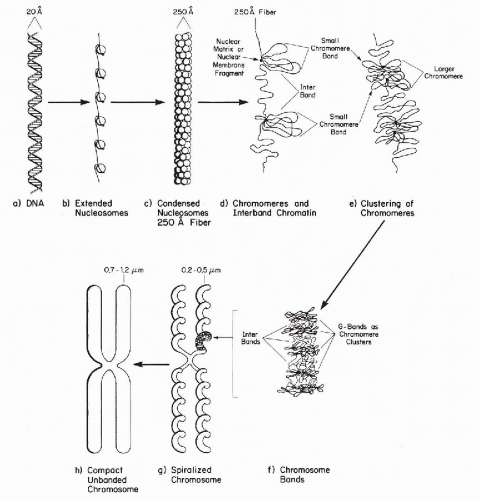
In suitable preparations, it is clear that each chromatid is in the form of a single helical coil, sometimes referred to as the major coil (see Fig. 4-2B). In some plant species, the strand making up the major coil is composed of smaller or minor coils (i.e., the chromatid is a coiled coil). It is to be noted that the minor coil is too large to be the Watson-Crick double helix of DNA, which may be found as fine strands at the next level of resolution. The chromosomal structure at metaphase would consist of two chromatids, each of which is coiled-coiled coil, the smallest coil being the DNA double helix. This is a useful model to keep in mind, but it may represent an oversimplification. Electron micrographs of whole human chromosomes at metaphase exhibit what has been called a folded fiber structure, in which the fibers appear sharply but randomly bent or angulated into meshwork (Fig. 4-3). These fibers, assuming there is a protein coat, are about the right dimensions for DNA molecules. The evidence appears to be consistent with the view that each chromatid represents a tangle of single-strand DNA, forming the Watson-Crick double helix (Fig. 4-4) (Dupraw, 1966; Miles, 1964; Bahr, 1977; Therman, 1993). There are several theories pertaining to the relationship of the primary DNA molecule to the organization of the chromosome and chromosomal banding. An example of this proposal by Comings is shown in Figure 4-5.
STAGES OF MITOSIS
Living things grow and maintain themselves in large measure because their cells are capable of multiplying by successive division. The steps observed in nuclear division are called mitosis or, more precisely, mitotic division. Although the stages of nuclear division are not sharply demarcated, they are conveniently referred to as:
prophase
prometaphase
metaphase
anaphase
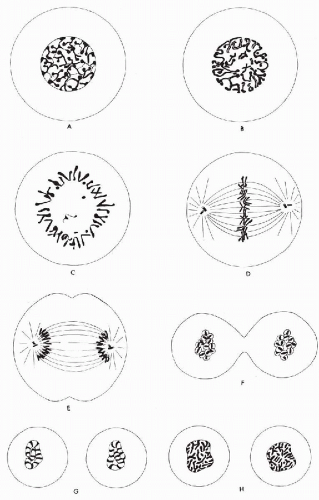
Mitosis is a complex process, which includes a break-down of the nuclear envelope, chromatin condensation, and chromosome segregation. A brief description of these stages will first be given to provide a framework for a more detailed discussion.
Prophase proceeds from the first visible signs of cell division until the breakdown of the nuclear envelope. During the prophase, the chromosomes have condensed and appear as long rod-like structures. Prometaphase starts with the disruption of the nuclear envelope. Metaphase is the period during which the chromosomes become aligned on the central metaphase plate. Anaphase begins with the abrupt separation of the chromatids into daughter chromosomes as they proceed toward opposite poles of the cell. Finally, the nuclear membrane becomes reconstituted during telophase.
Prophase
The transition from the G2 phase to the M phase of the cell cycle is not a sharply defined event. The chromatin, which is diffuse in interphase, slowly condenses into welldefined chromosomes, the exact number of which is a characteristic of the particular species; each chromosome has duplicated during the preceding S phase and consists of two sister chromatids joined at a specific point along their length by the centromere. While the chromosomes are condensing, the nucleolus begins to disassemble and gradually disappears.
Within the nucleus itself, the first sign of prophase is an accentuation of the chromocenters and a net-like pattern (Fig. 4-7B; see Fig. 4-6A). Several condensations of chromatin appear at the periphery of the nucleus, whence thin strands of chromatin extend into the center of the nucleus (see Fig. 4-7B). In females, the inactive X chromosome (Barr body) is larger than other chromocenters and is readily visible as a triangular condensation of chromatin (see Fig. 4-7). These strands and chromocenters are the condensing chromosomes. By this time, the chromosomes are probably doubled into the two chromatids or daughter chromosomes-to-be, but the double structure is sometimes difficult to visualize (Fig. 4-8A; see Fig. 4-6B). It is more evident in chromosomes that have been exposed to colchicine (a drug that inhibits mitosis) and which have been treated with hypotonic salt solutions. With the breakdown of the nuclear membrane, the chromosomes are quite distinct and are arranged into a circular position, known as a hollow spindle or prometaphase rosette (see below) (see Figs. 4-6C and 4-8B,C).
At the beginning of prophase, the cytoplasmic microtubules, which are part of the cytoskeleton (see Chap. 2), disassemble, forming a large pool of tubulin molecules. These molecules are then reused in the construction of the main component of the mitotic apparatus, the mitotic spindle. This is a bipolar fibrous structure, largely composed of microtubules, that assembles initially outside the nucleus. The focus for the spindle formation is marked in most animal cells by the centrioles (see Chap. 2). The cell’s original pair of centrioles replicates by a process that begins immediately before the S phase to give rise to two pairs of centrioles, which separate and travel to the opposite poles of the cell (see Fig. 4-6D). Each centriole pair now becomes part of a mitotic center that forms the focus for a radial array of microtubules, the aster (from Latin, aster = star). Initially, the two asters lie side by side, close to the nuclear envelope. By late prophase, the bundles of polar microtubules that interact between the two asters (seen as polar fibers in the light microscope) preferentially elongate and appear to push the two asters apart along the outer part of the nucleus. In this way, a bipolar mitotic spindle is formed.
Prometaphase
Prometaphase starts abruptly with the disruption of the nuclear envelope, which breaks up into membrane fragments that are indistinguishable from bits of endoplasmic reticulum (see Fig. 4-6C). These fragments remain visible around the spindle during mitosis. Specialized structures called kinetochores develop on either face of the centromeres and become attached to a special set of microtubules, called kinetochore fibers or kinetochore microtubules. These fibers radiate in opposite directions from the sides of each chromosome and interact with the fibers of the bipolar spindle. The chromosomes are thrown into agitated motion by the interactions of their kinetochore fibers with other components of the spindle.
Metaphase
In phase cinematography of living cells, the chromosomes may be seen to undergo slow to and fro writhing movements until they finally become aligned on an equatorial plane. This plane bisects the mitotic spindle. As a result of their prometaphase oscillations, arrangement of all the chromosomes is such that their centromeres lie in one plane. The kinetochore fibers seem to be responsible for aligning the chromosomes halfway between the spindle poles and for orienting them with their long axes at right angles to the spindle axis. Each chromosome is held in tension at the metaphase plate by the paired kinetochores, with their associated fibers pointing to opposite poles of the spindle (see Figs. 4-6D and 4-8D).
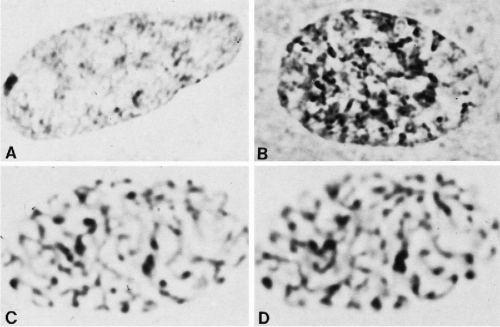 Figure 4-7 Interphase appearance and stages of prophase condensation. A. Interphase nucleus with sex chromatin body. Note the network of fine chromatin threads. B. An early stage of prophase showing accentuation of the chromatin network and of the peripheral chromocenters. (C,D) A somewhat later stage of prophase. The same nucleus photographed at two focal levels. (A,B, × 3,900; C,D, ×4,350). (From Miles CP. Chromatin elements, nuclear morphology and midbody in human mitosis. Acta Cytol 8:356-363, 1964.) |
Anaphase
The metaphase may last for several hours. As if triggered by a special signal, anaphase begins abruptly as the paired kinetochores on each chromosome separate, allowing each chromatid to be pulled slowly toward a spindle pole (see Fig. 4-6E). All chromatids are moved toward the pole they face at the same speed. During these movements, kinetochore fibers shorten as the chromosomes approach the poles. At about the same time, the spindle fibers elongate and the two poles of the polar spindle move farther apart. Soon after separation, the chromosomes appear at both poles as dark-staining masses (see Fig. 4-8E). The anaphase stage typically lasts only a few minutes.
In the meantime, the cell has become elongated, and a constriction furrow begins to appear at the level of the metaphase equator (see Figs. 4-9A and Fig. 4-6F). This process of cytoplasmic division is called cytokinesis. Although cytokinesis usually follows chromosomal division, the two processes are not necessarily dependent on one another. Chromosomal division may occur without cytokinesis (thereby producing a cell with double the normal complement of chromosomes). Less commonly, in some lower species, anucleated cytoplasm may undergo successive divisions.
The constriction furrow extends between the two daughter cells until only a narrow strand of cytoplasm is left. At this point, a distinct granule, the midbody, may sometimes be seen at the narrowest part of the cytoplasmic strand (see Fig. 4-9B). The midbody is formed, at least in part, by the spindle fibers compressed into a tight bundle. The precise significance and fate of the midbody are not known.
Telophase
Some details of telophase are worthy of attention. In late anaphase, after or during cytokinesis, the compact mass of chromosomes begins to swell. In optimal material, each chromosome appears to form a distinct small vesicle, possibly by inducing the formation of a proprietary segment of the nuclear membrane, as suggested by Koss (1998) (Fig. 4-10; see Fig. 4-6F). In abnormal divisions, the process may sometimes end at this stage, with the cell thus containing numerous micronuclei (Fig. 4-11). Normally, the vesicles seem to fuse rapidly together to form a convoluted tubule (see Figs. 4-12 and 4-6G). Probably the vesicle and tubule membranes break down at points of contact so that, ultimately, a continuous nuclear membrane is formed around both groups of daughter chromatids.
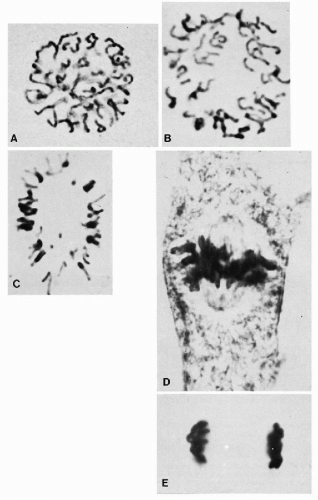 Figure 4-8 A. Late prophase, just before the breakdown of the nuclear membrane. The double structure can be visualized in some of these chromosomes. B. Nearing metaphase, the chromosomes show further contraction and (C) tend to congregate toward the periphery of the figure (“hollow spindle” arrangement). D. Chromosomes aligned on the metaphase plate. Note the spindle fibers converging on the centrioles. E. Late anaphase groups of daughter chromosomes. (A, × 2,220; B, × 3,450; C,D, × 3,150; E, ×3,120; D,E, phase contrast.) (From Miles, CP. Chromatin elements, nuclear morphology and midbody in human mitosis. Acta Cytol 8:356-363, 1964.) |
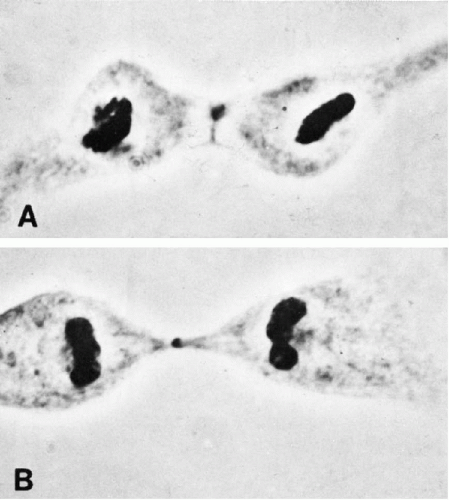 Figure 4-9 A. Division of the cytoplasm (cytokinesis). B. Later stage of cytokinesis. The midbody is the small central granule (phase contrast × 1,560). (From Miles, CP. Chromatin elements, nuclear morphology and midbody in human mitosis. Acta Cytol 8:356-363, 1964.) |
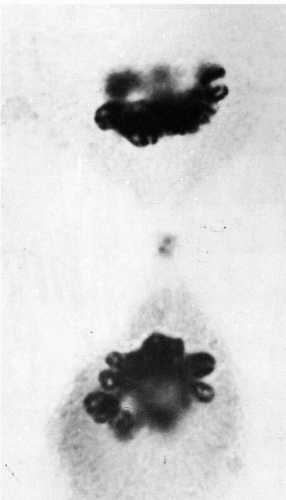 Figure 4-10 Beginning of telophase reconstruction. Each chromosome appears to form a small vesicle. The dark double structure at the center of the spindle conceivably represents a divided midbody (phase contrast × 2,250). (From Miles, CP. Chromatin elements, nuclear morphology and midbody in human mitosis. Acta Cytol 8:356-363, 1964.) |
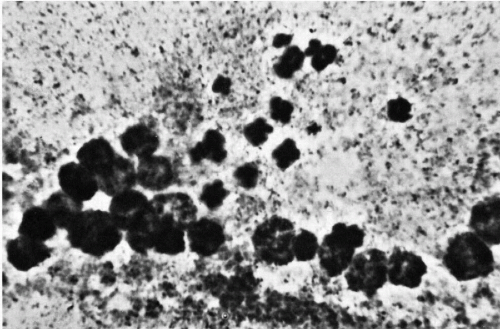 Figure 4-11 Abnormal mitosis with micronuclei, presumably formed through failure of chromosomal vesicles to coalesce (colchicine-treated culture; aceto-orcein stain ×1,610). (From Miles, CP. Chromatin elements, nuclear morphology and midbody in human mitosis. Acta Cytol 8:356-363, 1964.) |
The elongating chromosomes now appear at right angles to the tubule walls, thus to some extent, mimicking prophase appearances (see Figs. 4-13, 4-6H, and 4-12B). The outline of the nucleus gradually becomes less convoluted, and nucleolar material appears at the inner edges of the nucleus. The reticular appearance of the telophase nucleus (Fig. 4-14) gradually fades into the less-distinct interphase pattern.
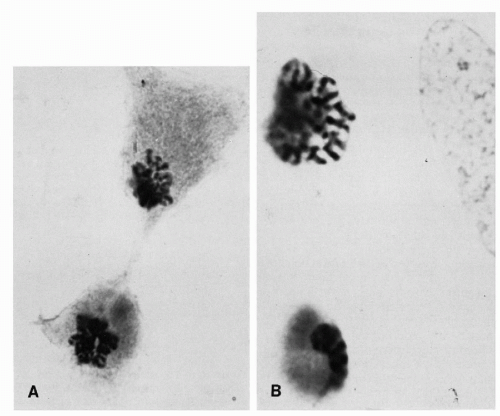 Figure 4-12 Vesicles coalescing into tubules. A. Note chromosomes arranged at right angles to the long axis of the tubule (aceto-orcein stain × 1,120; B, 1,610). (From Miles, CP. Chromatin elements, nuclear morphology and midbody in human mitosis. Acta Cytol 8:356-363, 1964.) |
As the separated daughter chromatids arrive at the poles, the kinetochore fibers disappear. The polar fibers elongate still farther, the condensed chromatin expands once more, the nucleoli begin to reappear, and the mitosis comes to an end.
Cytokinesis
As described above, the cytoplasm divides by a process known as cleavage, which usually starts sometime during late anaphase or telophase. The membrane around the middle of
the cell, perpendicular to the spindle axis and between the daughter nuclei, is drawn inward to form a cleavage furrow, which gradually deepens until it encounters the remains of the mitotic spindle between the two nuclei (see Figs. 4-6F and 4-9). This narrow bridge, which contains a dark granule, the midbody, may persist for some time before it finally breaks at each end, leaving two completed, separated daughter cells (Miles, 1979; Alberts, 1983; Brachet, 1985; Levitan, 1988; Edlin, 1990; Therman, 1993).
the cell, perpendicular to the spindle axis and between the daughter nuclei, is drawn inward to form a cleavage furrow, which gradually deepens until it encounters the remains of the mitotic spindle between the two nuclei (see Figs. 4-6F and 4-9). This narrow bridge, which contains a dark granule, the midbody, may persist for some time before it finally breaks at each end, leaving two completed, separated daughter cells (Miles, 1979; Alberts, 1983; Brachet, 1985; Levitan, 1988; Edlin, 1990; Therman, 1993).
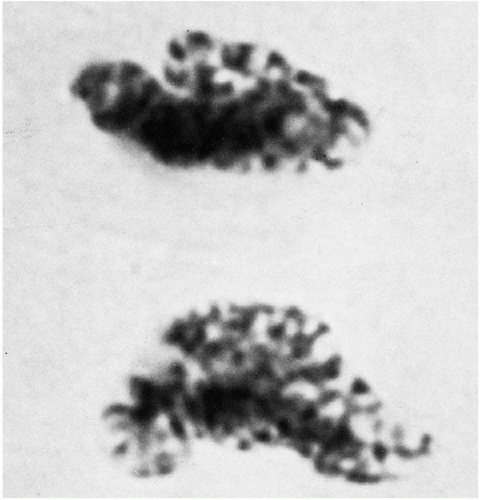 Figure 4-13 Late stages of telophase beginning to mimic prophase appearance. One daughter nucleus still shows tubule structure (aceto-orcein stain, ×3,150). (From Miles, CP. Chromatin elements, nuclear morphology and midbody in human mitosis. Acta Cytol 8:356-363, 1964.) |
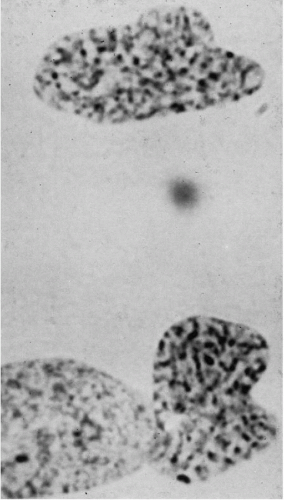 Figure 4-14 Telophase. The daughter nuclei still appear somewhat convoluted, but no suggestion of tubule remains. (Part of an interphase nucleus impinges on one daughter nucleus.) Some of the spoke-like chromosomal elements appear distinctly double, as does the larger bipartite chromocenter in one nucleus (the sex chromatin body?) (aceto-orcein stain × 2,520). (From Miles, CP. Chromatin elements, nuclear morphology and midbody in human mitosis. Acta Cytol 8:356-363, 1964.) |
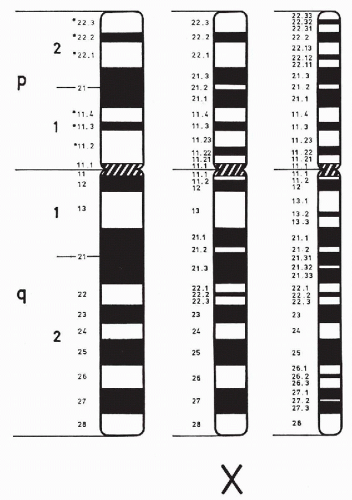
THE NORMAL HUMAN CHROMOSOME COMPLEMENT
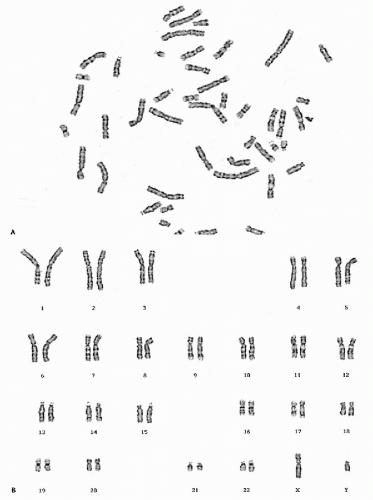
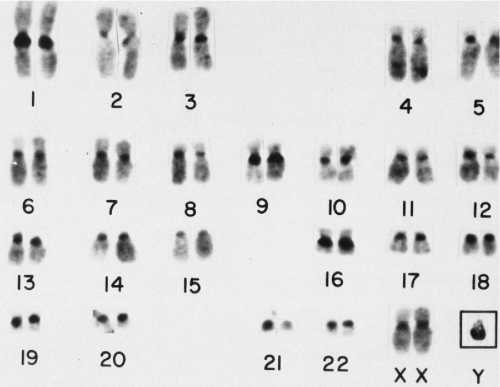
Before 1956, the number of human chromosomes was believed to be 48, and the XX-XY mechanism of sex determination was assumed to work in the same way as it does in the fruit fly, Drosophila. Both of these notions about human chromosomes were eventually proved wrong. The year 1956 is often given as the beginning of modern human cytogenetics; indeed, the discovery by Tjio and Levan in 1956 that the human chromosome number is 46 (Fig. 4-15), and not 48, was the starting point for subsequent spectacular developments in human cytogenetics. Contemporary techniques (use of colchecine/colcemid, culture methodologies, hypotonic treatment) confirmed that normal human cells have 46 chromosomes, two sex chromosomes (X,X or X,Y) and 44 autosomes. These can be seen and classified in the metaphase stage of cell division.
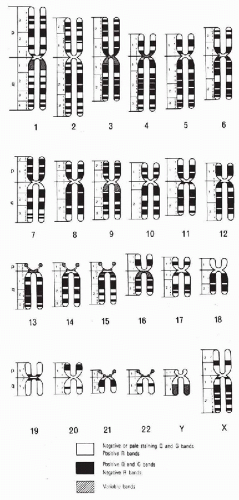
In 1970, Caspersson and his colleagues applied fluorescence microscopy, which they had originally used to study plant chromosomes, to the analysis of the human karyotype. They discovered that the chromosomes consist of differentially fluorescent cross bands of various lengths. Careful study of these bands made possible the identification of all human chromosomes. This discovery was followed by a host of different banding techniques. The most commonly employed technique is trypsin or G-banding (see Fig. 4-15). Chromosome preparations are pretreated with trypsin before staining them with Giemsa stain (hence, Giemsa or G-banding). By means of such banding, each chromosome (homologue) can be identified by the resulting alternating light and dark band patterns specific to that particular chromosome. Another banding procedure, which gives only slightly different results, involves staining with a fluorescent dye, quinacrine dihydrochloride, which thus yields quinacrine or Q-bands. These bands fluoresce under ultraviolet light with varying degrees of brightness, similar to the light and dark bands produced by G-banding. The banding of elongated prophase or prometaphase chromosomes makes it possible to define chromosome segments and breakpoints even more accurately (Bergsma, 1972; Yunis, 1974; Hsu, 1979; Emery and Rimoin, 1983; Mange and Mange, 1990). The C-banding technique is used to highlight the constitutive chromatin region of the chromosomes, usually the centromeres and the long arm of chromosome Y. The chromosomal preparations are exposed to barium hydroxide-saturated solution and stained with Giemsa.
With the exception of the sex chromosomes X and Y, the chromosomes occur in pairs, each pair composed of two identical chromosomes or homologues (from Greek, homo = same). Each pair of chromosomes has been numbered from 1 to 22 in order of length. The pairs are further divided into seven subgroups designated 1-3, 4-5, 6-12, 13-15, 16-18, 19-20, 21-22, or by letter A, B, C, D, E, F, and G, respectively (Fig. 4-16; see Fig. 4-15). The centromere, or primary constriction, is in a constant position on any given chromosome. In the terminology commonly employed for human chromosomes, the chromosome is metacentric if the centromere is located at the center of the chromosome, thus making both arms equal in length; a chromosome is submetacentric if one arm is longer than the other; a chromosome is acrocentric (acro = end) or subtelocentric if the centromere is located very close to the end of the short arm.
The pairs of chromosomes at metaphase can be accurately classified into the seven groups by using the characteristics of length and centromere position. The two groups of acrocentric chromosomes, D (13-15) and G (21-22), for example, are easily identifiable, especially in colchicine-treated preparations. Colchicine prevents (among other effects) the centromere from dividing but does not interfere with chromatid separation. Thus, the acrocentrics remain joined at one end and come to resemble a wishbone or an old-fashioned clothespin. However, distinguishing chromosomes within groups was difficult and, sometimes impossible, until banding techniques were discovered.
Additions or deletions of portions of the chromosomes are designated by chromosome numbers and band numbers followed by p or q and + or − signs. In this manner, a precise identification of chromosomal segments, which are missing, added, or translocated can be achieved. Specialized nomenclature has been established to denote changes in chromosome number and structure (ISCN, 1995).
The ability to identify every chromosome by number has led to a slight change in the rule correlating number with chromosomal length. It has been found that the chromosome that accounts for Down’s syndrome (see below) is, in fact, the shortest and not the next-to-shortest chromosome. However, to preserve the synonym trisomy 21 for Down’s syndrome, the shortest chromosome is designated as 21 and the nexts hortest chromosome is designated as 22.
Certain other chromosomal features, although not of great importance in identifying particular homologues, may ultimately be of significance in the study of pathology. The long and short acrocentrics (13-15 and 21-22 groups) often exhibit a small structure on the short arms, called a satellite. Satellites, when well visualized, consist of short, thin filaments surmounted by a tiny mass of chromatin. Satellites are close to the limits of resolution, and they can rarely be observed on all the acrocentrics within one cell. Failure to demonstrate them is probably due to technical difficulties. It is known, though, that some individuals show very conspicuous satellites, although, once again, not on all of the acrocentrics; there has been no convincing evidence that these larger satellites are related to any disease state.
Individual or familial differences may also be observed in the size and centromere position of chromosomes in normal persons. Size differences were first clearly shown for the Y chromosome (Sandberg, 1985a, 1985b).
In addition to cytogenetic techniques for identifying individual chromosomes and their bands, sub-bands, and structures (Fig. 4-17), techniques have been developed recently for identifying chromosomes based on unique DNA sequences within each chromosome. This approach allows the recognition of specific chromosomes, or their parts, in interphase nuclei, thus dispensing with the more laborious process of metaphase preparation, or in situations when metaphases cannot be obtained. Fluorescent in situ hybridization (FISH) with molecular “paint” probes to specific chromosomes and their components has become an established laboratory technique (see Fig. 2-31). It allows the analysis of cells and tissues for the presence of chromosomal abnormalities. However, detailed karyotype analysis still requires optimal metaphases for their construction (Cannizzaro and Shi, 1997; Montgomery et al, 1997).
Heterochromatin
Another feature of chromosomes that shows familial differences, probably unrelated to disease, is the secondary constriction. (The primary constriction is at the centromere where the spindle fibers attach during mitosis; see earlier.) Readily visible in the microscope are secondary constrictions in the long arms near the centromere on chromosomes 1, 9, and 16. In normal cells, these constrictions are seen only occasionally and seldom in more than one homologue in a given cell. These constrictions are usually observed near centromeric sites on most chromosomes. At these sites, most chromosomes have small blocks of chromatin that replicate their DNA after the other chromosomal segments have completed DNA synthesis (e.g., late-labeling DNA). Such sites can also be selectively stained with the C-banding technique, centromeric heterochromatic stain (Fig. 4-18). In many species, such dark-staining, late-labeling segments are referred to as heterochromatin. In some species, these segments do not decondense in the interphase nucleus but rather remain as dark-staining masses of chromatin called chromocenters. In general, such heterochromatin segments are genetically inert (do not contain functioning genes and do not synthesize RNA). They are believed to have something to do with maintaining the structure of the chromosome; the material, therefore, is called constitutive heterochromatin. The latter is differentiated from facultative heterochromatin, which is condensed in some cells and not in others and, in contrast to constitutive heterochromatin, reflects some of the stable differences in genetic activity adopted by different cell types (e.g., embryonic cells seemingly contain very little, and some highly specialized cells contain a great deal of heterochromatin). Facultative heterochromatin is not known to contain the large number of highly repeated DNA sequences (satellite DNAs), which is characteristic of constitutive heterochromatin (Bahr, 1977; Lima-de-Faria, 1983; Therman, 1993). Although chromocenters (except for the sex chromatin body; see below) may vary in their appearance in the nuclei of human cells and, in some, they are difficult to visualize, only polymorphonuclear leukocytes are an exception. In the nuclear lobes of these cells, the constitutive heterochromatin of chromosome 1 and, perhaps other chromosomes, is observed as a peripheral chromocenter. With a somewhat similar technique, the C-band heterochromatin of chromosome 9 can be identified in interphase nuclei of lymphocytes.
GERM CELL FORMATION, MEIOSIS, AND SEX DETERMINATION
Germ Cell Formation
As has been stated, most human cells contain 46 chromosomes. The germ cells, sperm and ovum, constitute an important exception. Since the individual develops from the union of sperm and ovum, to preserve the proper somatic number of 46, these cells can have only 23 chromosomes each. Thus, the developing germ cell must lose half its chromosomes. The product of the union of the spermatozoon and the ovum, or the zygote, will then receive 23 chromosomes from the mother and 23 from the father. A type of cell division known as meiosis fulfills these requirements (Fig. 4-19).
It is clear that normal development will require that the zygote receive a set of similar chromosomes (e.g., one No. 1, one No. 2, and so on) from each parent. A set of 23 maternal or paternal chromosomes is a haploid set, and the final two sets of homologues form a diploid set.
Meiosis
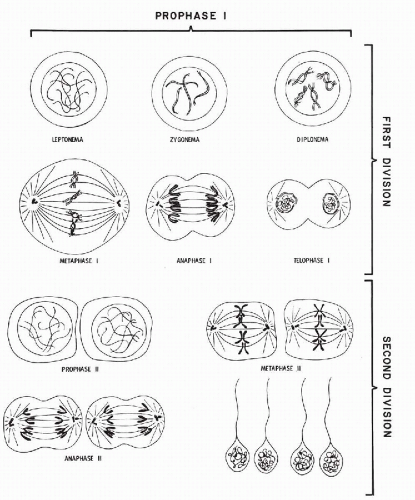
The fundamental mechanism of meiosis serves to ensure that each germ cell acquires a precise set of 23 homologues, including either an X or a Y chromosome. Meiosis essentially consists of two separate divisions, referred to as the first and the second meiotic divisions (see Fig. 4-19).
First Meiotic Division
The prophase sequence of this division has been divided into several stages named leptonema, zygonema, pachynema, diplonema, and diakinesis. The chromosomes in leptonema (from Greek, lepto = thin and nema = thread) condense out as long convoluted threads. In zygonema (from Latin, zygo = pair), homologous chromosomes come together and pair, point for point, along their lengths. This process is called synapsis of the homologues, and the closely aligned synapsed pair is called a bivalent.
At the beginning of pachynema (from Greek, pachy = thick), pairing is complete, and the chromosomes become shorter and thicker. By this stage, each homologue may appear doubled into its two chromatids; hence, four units are seen, and the bivalent has become a tetrad. In diplonema (from Latin, diplo = double), the homologues begin to move away from one another, but they usually continue to remain joined at one or more points along their lengths. The involved segments near such points will resemble an X, or a cross, hence the name chiasma (plural, chiasmata) for such points.
In diakinesis, the tetrad continues to loosen, until at the
first meiotic metaphase, the homologues separate completely and pass to opposite poles.
first meiotic metaphase, the homologues separate completely and pass to opposite poles.
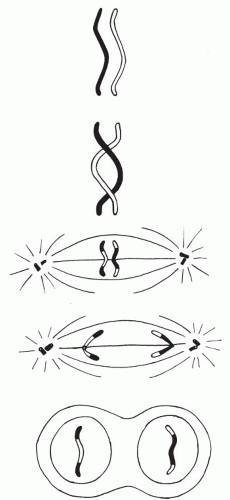
Crossover
The appearance of chiasmata is associated with the reciprocal exchange of segments of homologous chromatids that had tightly synapsed together in the earlier stage. Perhaps this process can be more readily grasped if we visualize the paternal homologue as a single column of soldiers that becomes aligned with a similar column, the homologue of maternal origin. If a few of the soldiers simply exchange places with an equal number from the opposite group, the composition of each column becomes completely different, but the general appearance of the column remains unchanged. In genetic terms, a crossover has occurred, and each column now represents a new combination of soldiers (Fig. 4-20).
Chromosomal segments cannot be exchanged quite so readily as soldiers in a column, since breaks in the chromosomes are probably necessary, and each break apparently prevents the occurrence of a similar break in the near vicinity. Consequently, the synapsed chromosomes seldom exchange more than one or two segments. Thus, the final germ cell does not necessarily receive unaltered paternal or maternal homologues. Many of its chromosomes will consist of rejoined segments from both parents. Although the behavior of the X chromosome in female meiosis is similar to that of the autosomes, the behavior of the X and Y in male meiosis is an exception to the rule. The X and Y chromosomes in the developing spermatocyte do not synapse together and, consequently, do not exchange segments by crossing over. Instead, the human X and Y chromosomes pair at the distal
ends of their short arms during male meiosis. There is formation of a synaptonemal complex between X and Y chromosomes in this region. Recent molecular studies have shown that there is DNA homology between X and Y chromosomes at their distal short arms, where there is a single obligatory crossing over between X and Y during meiosis. As a result, loci mapping in this region do not show strict sex linkage; accordingly, this homologous segment of the X and Y chromosomes is referred to as the pseudoautosomal region (Sandberg, 1983a).
ends of their short arms during male meiosis. There is formation of a synaptonemal complex between X and Y chromosomes in this region. Recent molecular studies have shown that there is DNA homology between X and Y chromosomes at their distal short arms, where there is a single obligatory crossing over between X and Y during meiosis. As a result, loci mapping in this region do not show strict sex linkage; accordingly, this homologous segment of the X and Y chromosomes is referred to as the pseudoautosomal region (Sandberg, 1983a).
Second Meiotic Division
The second meiotic division is much more akin to a somatic or mitotic division, with separation of the chromatids. All of the resulting daughter cells are haploid, that is, contain 23 chromosomes. As a result of crossing over in meiosis I, the genetic content of each haploid cell is a mixture of paternal and maternal genes.
Meiosis not only serves the fundamental need of reducing the chromosomal number of the germ cells but also constitutes a kind of lottery that vastly increases the possibilities for genetic variation. Not only does each germ cell draw at random one or the other homologue, but these homologues may themselves have already been altered through reciprocal exchange of segments. Meiosis is the principal reason for the enormous diversity, even among members of the same family (Roberts and Pembrey, 1985).
Sex Determination
The sex chromosomes, the male Y and the female X, differ from the nonsex chromosomes or autosomes. Whereas the female has two X chromosomes, the male has only one X and one Y. During the process of meiosis, each germ cell ends up with a precise haploid set of autosomes. In sperm, each haploid set will also include either an X or a Y chromosome. Thus, by chance, roughly 50% of sperm will bear an X and 50% a Y. Since the mother has only one kind of sex chromosome, all of the ova contain a single X. If an ovum is fertilized by an X-bearing sperm, a female zygote will result (46,XX); if by a Y-bearing sperm, the offspring will be male (46,XY). Thus, it is the paternal chromosome that determines the sex of the child (Ohno et al, 1962




Stay updated, free articles. Join our Telegram channel

Full access? Get Clinical Tree