INTRODUCTION
Poxviruses are the largest and most complex of viruses infecting humans. The family encompasses a large group of agents that are similar morphologically and share a common nucleoprotein antigen. Infections with most poxviruses are characterized by a rash, although lesions induced by some members of the family are markedly proliferative. The group includes variola virus, the etiologic agent of smallpox—the viral disease that has affected humans throughout recorded history.
Even though smallpox was declared eradicated from the world (in 1980) after an intensive campaign coordinated by the World Health Organization, there is concern that the virus could be reintroduced as a biologic weapon. There is a continuing need to be familiar with vaccinia virus (used for smallpox vaccinations) and its possible complications in humans. It is also necessary to be aware of other poxvirus diseases that may resemble smallpox and must be differentiated from it by laboratory means. Lastly, vaccinia virus is under intensive study as a vector for introducing active immunizing genes as live-virus vaccines for a variety of viral diseases of humans and domestic animals.
PROPERTIES OF POXVIRUSES
Important properties of the poxviruses are listed in Table 34-1.
| Virion: Complex structure, oval or brick shaped, 300–400 nm in length × 230 nm in diameter; external surface shows ridges; contains core and lateral bodies |
| Composition: DNA (3%), protein (90%), lipid (5%) |
| Genome: Double-stranded DNA, linear; size, 130–375 kbp; has terminal loops; has low G + C content (30–40%) except for Parapoxvirus (63%) |
| Proteins: Virions contain more than 100 polypeptides; many enzymes are present in core, including transcriptional system |
| Envelope: Virion assembly involves formation of multiple membranes |
| Replication: Cytoplasmic factories |
| Outstanding characteristics: Large and complex viruses; very resistant to inactivation Virus-encoded proteins help evade host immune defense system Smallpox was the first viral disease eradicated from the world |
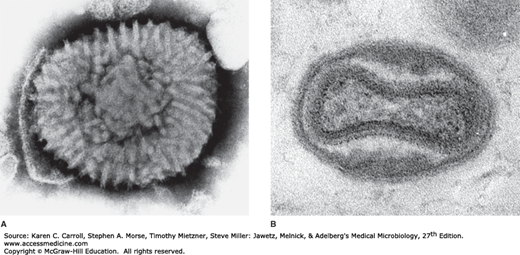
Poxviruses are large enough to be seen as featureless particles by light microscopy. By electron microscopy, they appear to be brick-shaped or ellipsoid particles measuring about (300–400) × 230 nm. Their structure is complex and conforms to neither icosahedral nor helical symmetry. The external surface of particles contains ridges. An outer lipoprotein membrane, or envelope, encloses a core and two structures of unknown function called lateral bodies (Figure 34-1).
FIGURE 34-1
Electron micrographs of vaccinia (Orthopoxvirus) virions. A: Negatively stained particle showing ridges or tubular elements covering the surface (228,000×). (Reproduced with permission from Dales S: The uptake and development of vaccinia virus in strain L cells followed with labeled viral deoxyribonucleic acid. J Cell Biol 1963;18:51.) B: Thin section of vaccinia virion showing a central biconcave core, two lateral bodies, and an outer membrane (220,000×). (Reproduced with permission from Pogo BGT, Dales S: Two deoxyribonuclease activities within purified vaccinia virus. Proc Natl Acad Sci USA 1969;63:820.)
The core contains the large viral genome of linear double-stranded DNA (130–375 kbp). The complete genomic sequence is known for several poxviruses, including vaccinia and variola. The vaccinia genome contains about 185 open reading frames. The DNA contains inverted terminal repeats of variable length, and the strands are connected at the ends by terminal hairpin loops. The inverted terminal repeats may include coding regions, so some genes are present at both ends of the genome. The DNA is rich in adenine and thymine bases.
The chemical composition of a poxvirus resembles that of a bacterium. Vaccinia virus is composed predominantly of protein (90%), lipid (5%), and DNA (3%). More than 100 structural polypeptides have been detected in virus particles. A number of the proteins are glycosylated or phosphorylated. The lipids are cholesterol and phospholipids.
The virion contains a multiplicity of enzymes, including a transcriptional system that can synthesize, polyadenylate, cap, and methylate viral mRNA.
Poxviruses are divided into two subfamilies based on whether they infect vertebrate or insect hosts. The vertebrate poxviruses fall into nine genera, with the members of a given genus displaying similar morphology and host range as well as some antigenic relatedness.
Most of the poxviruses that can cause disease in humans are contained in the genera Orthopoxvirus and Parapoxvirus; there are also several that are classified in the genera Yatapoxvirus and Molluscipoxvirus (Table 34-2).
| Genus | Virus | Primary Host | Disease |
|---|---|---|---|
| Orthopoxvirus | Variola (major and minor) | Humans | Smallpox (now eliminated) |
| Vaccinia | Humans | Localized lesion; used for smallpox vaccination | |
| Buffalopox | Water buffalo | Human infections rare; localized lesion | |
| Monkeypox | Rodents, monkeys | Human infections rare; generalized disease | |
| Cowpox | Cows | Human infections rare; localized ulcerating lesion | |
| Parapoxvirus | Orf | Sheep | Human infections rare; localized lesion |
| Pseudocowpox | Cows | ||
| Bovine papular stomatitis | Cows | ||
| Molluscipoxvirus | Molluscum contagiosum | Humans | Many benign skin nodules |
| Yatapoxvirus | Tanapox | Monkeys | Human infections rare; localized lesion |
| Yabapox | Monkeys | Human infections very rare and accidental; localized skin tumors |
The orthopoxviruses have a broad host range affecting several vertebrates. They include ectromelia (mousepox), camelpox, cowpox, monkeypox, vaccinia, and variola (smallpox) viruses. The last four are infectious for humans. Vaccinia virus differs only in minor morphologic respects from variola and cowpox viruses. It is the prototype of poxviruses in terms of structure and replication. Monkeypox can infect rodents, monkeys, and humans and may resemble smallpox clinically.
Some poxviruses have a restricted host range and infect only rabbits (fibroma and myxoma) or only birds. Others infect mainly sheep and goats (sheeppox, goatpox) or cattle (pseudocowpox, or milker’s nodule).
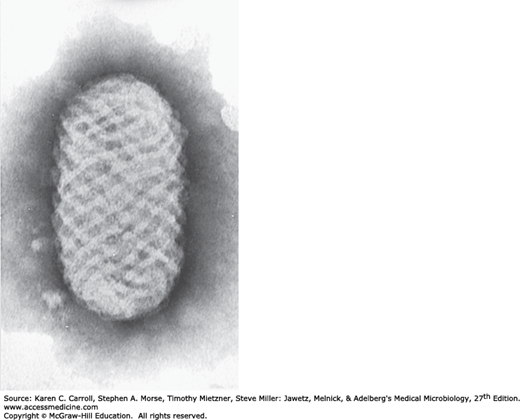
Parapoxviruses are morphologically distinctive. Compared with the orthopoxviruses, parapoxviruses are somewhat smaller particles (260 × 160 nm), and their surfaces exhibit a crisscross pattern (Figure 34-2). Their genomes are smaller (~135 kbp) and have a higher guanine plus cytosine content (63%) than those of the orthopoxviruses (170–250 kbp; G + C, 30–40%).
All vertebrate poxviruses share a common nucleoprotein antigen in the inner core. There is serologic cross-reactivity among viruses within a given genus but very limited reactivity across genera. Consequently, immunization with vaccinia virus affords no protection against disease induced by other genera of poxviruses.
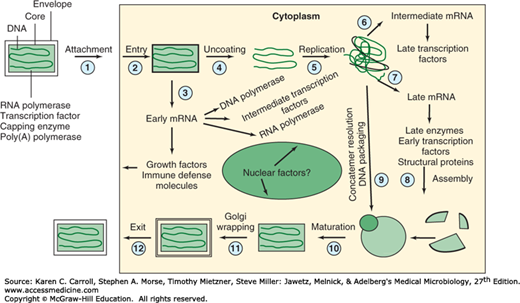
The replication cycle of vaccinia virus is summarized in Figure 34-3. Poxviruses are unique among DNA viruses in that the entire multiplication cycle takes place in the cytoplasm of infected cells. It is possible, however, that nuclear factors may be involved in transcription and virion assembly. Poxviruses are further distinguished from all other animal viruses by the fact that the uncoating step requires a newly synthesized, virus-encoded protein.
FIGURE 34-3
Outline of replication cycle of vaccinia virus. The replication of this large DNA virus occurs in the cell cytoplasm. (1) Virus particles attach to cells, (2) fuse with the cell membrane, and release viral cores into the cytoplasm. (3) The cores produce early mRNAs using viral enzymes and transcription factors contained within the cores; these mRNAs are translated into numerous viral proteins having replication functions. (4) The cores are uncoated and (5) viral DNA is replicated. (6 and 7) Intermediate and late genes are transcribed; products include viral structural proteins. (8–10) Assembly of infectious virions occurs on membrane structures. (11) Envelopes are acquired at the Golgi and plasma membrane, leading to (12) the release of enveloped progeny virions. (Reproduced with permission from Moss B: Poxviridae: The viruses and their replication. In Fields BN, Knipe DM, Howley PM [editors-in-chief]. Fields Virology, 3rd ed. Lippincott-Raven, 1996.)
Virus particles establish contact with the cell surface and fuse with the cell membrane. Some particles may appear within vacuoles. Viral cores are released into the cytoplasm. Among the several enzymes inside the poxvirus particle, there is a viral RNA polymerase that transcribes about half the viral genome into early mRNA. These mRNAs are transcribed within the viral core and are then released into the cytoplasm. Because the necessary enzymes are contained within the viral core, early transcription is not affected by inhibitors of protein synthesis. The “uncoating” protein that acts on the cores is among the more than 50 polypeptides made early after infection. The second-stage uncoating step liberates viral DNA from the cores; it requires both RNA and protein synthesis. The synthesis of host cell macromolecules is inhibited at this stage.
Poxviruses inactivated by heat can be reactivated either by viable poxviruses or by poxviruses inactivated by nitrogen mustards (which inactivate the DNA). This process is called nongenetic reactivation and is caused by the action of the uncoating protein. Heat-inactivated virus alone cannot cause second-stage uncoating because of the heat lability of the RNA polymerase. Apparently, the heat-killed virus provides the template and the second virus provides the enzymes needed for transcription. Any vertebrate poxvirus can reactivate any other vertebrate poxvirus.
Among the early proteins made after vaccinia virus infection are enzymes involved in DNA replication, including a DNA polymerase and thymidine kinase. Viral DNA replication occurs in the cytoplasm and appears to be accomplished by viral-coded enzymes. Viral DNA replication starts soon after the release of viral DNA in the second stage of uncoating. It occurs 2–6 hours after infection in discrete areas of the cytoplasm, which appear as “factories” or inclusion bodies in electron micrographs. The number of inclusion bodies per cell is proportionate to the multiplicity of infection, suggesting that each infectious particle can induce a “factory.” High rates of homologous recombination occur within poxvirus-infected cells. This has been exploited experimentally to construct and map mutations.
The pattern of viral gene expression changes markedly with the onset of replication of viral DNA. The synthesis of many of the early proteins is inhibited. There is a small intermediate class of genes whose expression temporally precedes the expression of the late class of genes. Late viral mRNA is translated into large amounts of structural proteins and small amounts of other viral proteins and enzymes.
The assembly of the virus particle from the manufactured components is a complex process. Some of the particles are released from the cell by budding, but the majority of poxvirus particles remain within the host cell. About 10,000 virus particles are produced per cell. How the multiple components of the transcription system are incorporated within the core of the assembling virus particle is unknown.
A polypeptide encoded by one of the early genes of vaccinia virus is closely related to epidermal growth factor and to transforming growth factor-α. Production of growth factors similar to epidermal growth factor by virus-infected cells could account for the proliferative diseases associated with members of the poxvirus family such as Shope fibroma (rabbits), Yaba tumor (monkeys), and molluscum contagiosum viruses (humans).
Several poxvirus genes resemble mammalian genes for proteins that would inhibit host defense mechanisms. Examples include tumor necrosis factor receptor, interferon-γ receptor, interleukin-1 receptor, and a complement-binding protein. These poxvirus-encoded host defense modifiers presumably counter the complement and cytokine networks important in the host immune response to viral infection, allowing enhanced virus replication and, perhaps, facilitating virus transmission.