Pharmacogenetics
Pharmacogenetics is the study of the genetic basis for variation in drug response. In this broadest sense, pharmacogenetics encompasses pharmacogenomics, which employs tools for surveying the entire genome to assess multigenic determinants of drug response. Individuals differ from each other approximately every 300-1000 nucleotides, with an estimated total of 10 million single nucleotide polymorphisms (SNPs; single base pair substitutions found at frequencies ≥1% in a population) and thousands of copy number variations in the genome. Identifying which of these variants or combinations of variants have functional consequence for drug effects is the task of modern pharmacogenetics.
IMPORTANCE OF PHARMACOGENETICS TO VARIABILITY IN DRUG RESPONSE
Drug response is considered to be a gene-by-environment phenotype. An individual’s response to a drug depends on the complex interplay between environmental factors (e.g., diet, age, infections, drugs, exercise level, occupation, exposure to toxins, tobacco, and alcohol use) and genetic factors (e.g., gender, variants of drug transporters, and drug metabolizing enzymes expressed). Variation in drug response therefore may be explained by variation in environmental and genetic factors, alone or in combination.
Drug metabolism is highly heritable, with genetic factors accounting for most of the variation in metabolic rates for many drugs.
Comparison of intra-twin vs inter-pair variability suggests that ~75-85% of the variability in pharmacokinetic half-lives for drugs that are eliminated by metabolism is heritable. Extended kindreds may be used to estimate heritability. Inter- vs. intra-family variability and relationships among members of a kindred are used to estimate heritability. Using this approach with lymphoblastoid cells, cytotoxicity from chemotherapeutic agents was shown to be heritable, with ~20-70% of the variability in sensitivity to 5-fluorouracil, cisplatin, docetaxel, and other anticancer agents estimated as inherited.
For “monogenic” phenotypic traits, it is often possible to predict phenotype based on genotype. Several genetic polymorphisms of drug metabolizing enzymes result in monogenic traits. Based on a retrospective study, 49% of adverse drug reactions were associated with drugs that are substrates for polymorphic drug metabolizing enzymes, a proportion larger than estimated for all drugs (22%) or for top-selling drugs (7%). Prospective genotype determinations may result in the ability to prevent adverse drug reactions. Defining multigenic contributors to drug response will be much more challenging. For some multigenic phenotypes, such as response to antihypertensives, the large numbers of candidate genes will necessitate a large patient sample size to produce the statistical power required to solve the “multigene” problem.
GENOMIC BASIS OF PHARMACOGENETICS
PHENOTYPE-DRIVEN TERMINOLOGY
A trait (e.g., CYP2D6 “poor metabolism”) is deemed autosomal recessive if the responsible gene is located on an autosome (i.e., it is not sex-linked) and a distinct phenotype is evident only with nonfunctional alleles on both the maternal and paternal chromosomes. An autosomal recessive trait does not appear in heterozygotes. A trait is deemed codominant if heterozygotes exhibit a phenotype that is intermediate to that of homozygotes for the common allele and homozygotes for the variant allele. With the advances in molecular characterization of polymorphisms and a genotype-to-phenotype approach, many polymorphic traits (e.g., CYP2C19 metabolism of drugs such as mephenytoin and omeprazole) are now recognized to exhibit some degree of codominance. Two major factors complicate the historical designation of recessive, codominant, and dominant traits. First, even within a single gene, a vast array of polymorphisms (promoter, coding, non-coding, completely inactivating, or modestly modifying) are possible. Each polymorphism may produce a different effect on gene function and therefore differentially affect a measured trait. Second, most traits (pharmacogenetic and otherwise) are multigenic, not monogenic. Thus, even if the designations of recessive, codominant, and dominant are informative for a given gene, their utility in describing the genetic variability that underlies variability in drug response phenotype is diminished, because variability is likely to be multigenic.
TYPES OF GENETIC VARIANTS
A polymorphism is a variation in the DNA sequence that is present at an allele frequency of 1% or greater in a population. Two major types of sequence variation have been associated with variation in human phenotype: single nucleotide polymorphisms (SNPs) and insertions/deletions (indels). In comparison to base pair substitutions, indels are much less frequent in the genome and are of particularly low frequency in coding regions of genes. Single base pair substitutions that are present at frequencies ≥ 1% in a population are termed SNPs and are present in the human genome at ~1 SNP every few hundred to a thousand base pairs.
SNPs in the coding region are termed cSNPs (coding SNPs), and are further classified as nonsynonymous (or missense) or synonymous (or sense). Coding nonsynonymous SNPs result in a nucleotide substitution that changes the amino acid codon (e.g., proline [CCG] to glutamine [CAG]), which could change protein structure, stability, substrate affinities, or introduce a stop codon. Coding synonymous SNPs do not change the amino acid codon, but may have functional consequences (transcript stability, splicing). Typically, substitutions of the third base pair, termed the wobble position, in a 3 base pair codon, such as the G to A substitution in proline (CCG → CCA), do not alter the encoded amino acid. Base pair substitutions that lead to a stop codon are termed nonsense mutations. In addition, ~10% of SNPs can have more than 2 possible alleles (e.g., a C can be replaced by either an A or G), so that the same polymorphic site can be associated with amino acid substitutions in some alleles but not others.
Synonymous polymorphisms have sometimes been found to contribute directly to a phenotypic trait. One of the most notable examples is a polymorphism in ABCB1, which encodes P-glycoprotein, an efflux pump that interacts with many clinically used drugs. The synonymous polymorphism, C3435T, is associated with various phenotypes and results in a change from a preferred codon for isoleucine to a less preferred codon. Presumably, the less preferred codon is translated at a slower rate, which apparently changes the folding of the protein, its insertion into the membrane, and its interaction with drugs.
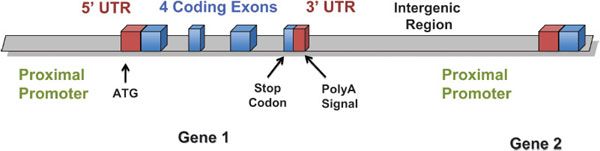
Noncoding SNPs may be in promoters, introns, or other regulatory regions that may affect transcription factor binding, enhancers, transcript stability, or splicing. Polymorphisms in noncoding regions of genes may occur in the 3′ and 5′ untranslated regions, in promoter or enhancer regions, in intronic regions, or in large regions between genes, intergenic regions (for nomenclature guide, see Figure 7–1). Noncoding SNPs in promoters or enhancers may alter cis– or trans-acting elements that regulate gene transcription or transcript stability. Noncoding SNPs in introns or exons may create alternative exon splicing sites, and the altered transcript may have fewer or more exons, or shorter or larger exons, than the wild-type transcript. Introduction or deletion of exonic sequence can cause a frame shift in the translated protein and thereby change protein structure or function, or result in an early stop codon, which makes an unstable or nonfunctional protein. Because 95% of the genome is intergenic, most polymorphisms are unlikely to directly affect the encoded transcript or protein. However, intergenic polymorphisms may have biological consequences by affecting DNA tertiary structure, interaction with chromatin and topoisomerases, or DNA replication. Thus, intergenic polymorphisms cannot be assumed to be without pharmacogenetic importance.
Figure 7–1 Nomenclature of genomic regions.
The second major type of polymorphism is indels. SNP indels can have any of the same effects as SNP substitutions: short repeats in the promoter (which can affect transcript amount), or insertions/deletions that add or subtract amino acids. A remarkable diversity of indels is tolerated as germline polymorphisms. A common glutathione-S-transferase M1 (GSTM1) polymorphism is caused by a 50-kilobase (kb) germline deletion, and the null allele has a population frequency of 0.3-0.5. Biochemical studies indicate that livers from homozygous null individuals have only ~50% of the glutathione conjugating capacity of those with at least 1 copy of the GSTM1 gene. The number of TA repeats in the UGT1A1 promoter affects the quantitative expression of this crucial glucuronosyltransferase in liver; 6 or 7 repeats constitute the most common alleles.
Some deletion and duplication polymorphisms can be seen as a special case of copy number variations (CNVs). Copy number variations involve large segments of genomic DNA that may involve gene duplications (stably transmitted inherited germline gene replication that causes increased protein expression and activity), gene deletions that result in the complete lack of protein production, or inversions of genes that may disrupt gene function. CNVs range in size from 1 kb to many megabases. CNVs appear to occur in ~10% of the human genome and in 1 study accounted for ~18% of the detected genetic variation in expression of around 15,000 genes in lymphoblastoid cell lines. There are notable examples of CNVs in pharmacogenetics; gene duplications of CYP2D6 are associated with an ultra-rapid metabolizer phenotype.
A haplotype, which is defined as a series of alleles found at a linked locus on a chromosome, specifies the DNA sequence variation in a gene or a gene region on 1 chromosome. For example, consider 2 SNPs in ABCB1, which encodes for the multidrug resistance protein, P-glycoprotein. One SNP is a T-to-A base pair substitution at position 3421 and the other is a C-to-T change at position 3435. Possible haplotypes would be T3421C3435, T3421T3435, A3421C3435, and A3421T3435. For any gene, individuals will have 2 haplotypes, 1 maternal and 1 paternal in origin. A haplotype represents the constellation of variants that occur together for the gene on each chromosome. In some cases, this constellation of variants, rather than the individual variant or allele, may be functionally important. In others, however, a single mutation may be functionally important regardless of other linked variants within the haplotype(s).
Two terms describe the relationship of genotypes at 2 loci: linkage equilibrium and linkage disequilibrium. Linkage equilibrium occurs when the genotype present at 1 locus is independent of the genotype at the second locus. Linkage disequilibrium occurs when the genotypes at the 2 loci are not independent of one another. In complete linkage disequilibrium, genotypes at 2 loci always occur together. Patterns of linkage disequilibrium are population specific and as recombination occurs linkage disequilibrium between 2 alleles will decay and linkage equilibrium will result.
ETHNIC DIVERSITY. Polymorphisms differ in their frequencies within human populations and have been classified as either cosmopolitan or population (or race and ethnic) specific. Cosmopolitan polymorphisms are those polymorphisms present in all ethnic groups, although frequencies may differ among ethnic groups. Likely to have arisen before migrations of humans from Africa, cosmopolitan polymorphisms are generally older than population-specific polymorphisms. The presence of ethnic and race-specific polymorphisms is consistent with geographical isolation of human populations. These polymorphisms probably arose in isolated populations and then reached a certain frequency because they are advantageous (positive selection) or, more likely, neutral to a population. African Americans have the highest number of population-specific polymorphisms in comparison to European Americans, Mexican Americans, and Asian Americans.
PHARMACOGENETIC STUDY DESIGN CONSIDERATIONS
PHARMACOGENETIC TRAITS
A pharmacogenetic trait is any measurable or discernible trait associated with a drug. Thus, enzyme activity, drug or metabolite levels in plasma or urine, blood pressure or lipid lowering produced by a drug, and drug-induced gene expression patterns are examples of pharmacogenetic traits. Directly measuring a trait (e.g., enzyme activity) has the advantage that the net effect of the contributions of all genes that influence the trait is reflected in the phenotypic measure. However, it has the disadvantage that it is also reflective of nongenetic influences (e.g., diet, drug interactions, diurnal, or hormonal fluctuation) and thus, may be “unstable.”
Stay updated, free articles. Join our Telegram channel

Full access? Get Clinical Tree