INTRODUCTION
Respiratory illnesses are responsible for more than half of all acute illnesses each year in the United States. The Orthomyxoviridae (influenza viruses) are a major determinant of morbidity and mortality caused by respiratory disease, and outbreaks of infection sometimes occur in worldwide epidemics. Influenza has been responsible for millions of deaths worldwide. Mutability and high frequency of genetic reassortment and resultant antigenic changes in the viral surface glycoproteins make influenza viruses formidable challenges for control efforts. Influenza type A is antigenically highly variable and is responsible for most cases of epidemic influenza. Influenza type B may exhibit antigenic changes and sometimes causes epidemics. Influenza type C is antigenically stable and causes only mild illness in immunocompetent individuals.
PROPERTIES OF ORTHOMYXOVIRUSES
Three immunologic types of influenza viruses are known, designated A, B, and C. Whereas antigenic changes continually occur within the type A group of influenza viruses and to a lesser degree in the type B group, type C appears to be antigenically stable. Influenza A strains are also known for aquatic birds, chickens, ducks, pigs, horses, and seals. Some of the strains isolated from animals are antigenically similar to strains circulating in the human population.
The following descriptions are based on influenza virus type A, the best-characterized type (Table 39-1).
| Virion: Spherical, pleomorphic, 80–120 nm in diameter (helical nucleocapsid, 9 nm) |
| Composition: RNA (1%), protein (73%), lipid (20%), carbohydrate (6%) |
| Genome: Single-stranded RNA, segmented (eight molecules), negative-sense, 13.6 kb overall size |
| Proteins: Nine structural proteins, one nonstructural |
| Envelope: Contains viral hemagglutinin and neuraminidase proteins |
| Replication: Nuclear transcription; capped 5′ termini of cellular RNA scavenged as primers; particles mature by budding from plasma membrane |
Outstanding characteristics: Genetic reassortment common among members of the same genus Influenza viruses cause worldwide epidemics |
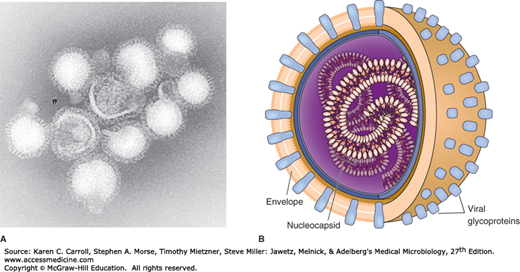
Influenza virus particles are usually spherical and about 100 nm in diameter (80–120 nm), although virions may display great variation in size (Figure 39-1).
FIGURE 39-1
Influenza virus. A: Electron micrograph of influenza virus A/Hong Kong/1/68(H3N2). Note the pleomorphic shapes and glycoprotein projections covering particle surfaces (315,000×). (Courtesy of FA Murphy and EL Palmer.) B: Schematic view of influenza. Virus particles have segmented genomes consisting of seven or eight different RNA molecules, each coated by capsid proteins and forming helical nucleocapsids. Viral glycoproteins (hemagglutinin and neuraminidase) protrude as spikes through the lipid envelope. (Reproduced with permission from Willey JM, Sherwood LM, Woolverton CJ: Prescott, Harley, and Klein’s Microbiology, 7th ed. McGraw Hill, 2008. © The McGraw-Hill Companies, Inc.)
The single-stranded, negative-sense RNA genomes of influenza A and B viruses occur as eight separate segments; influenza C viruses contain seven segments of RNA, lacking a neuraminidase gene. Sizes and protein-coding assignments are known for all the segments (Table 39-2). Most of the segments code for a single protein. The first 12–13 nucleotides at each end of each genomic segment are conserved among all eight RNA segments; these sequences are important in viral transcription.
| Genome Segment | Encoded Polypeptide | ||||
|---|---|---|---|---|---|
| Numbera | Size (Number of Nucleotides) | Designation | Predicted Molecular Weightb | Approximate Number of Molecules per Virion | Function |
| 1 | 2341 | PB2 | 85,700 | 30–60 | RNA transcriptase components |
| 2 | 2341 | PB1 | 86,500 | ||
| 3 | 2233 | PA | 84,200 | ||
| 4 | 1778 | HA | 61,500 | 500 | Hemagglutinin; trimer; envelope glycoprotein; mediates virus attachment to cells; activated by cleavage; fusion activity at acid pH |
| 5 | 1565 | NP | 56,100 | 1000 | Associated with RNA and polymerase proteins; helical structure; nucleocapsid |
| 6 | 1413 | NA | 50,000 | 100 | Neuraminidase; tetramer; envelope glycoprotein; enzyme |
| 7 | 1027 | M1 | 27,800 | 3000 | Matrix protein; major component of virion; lines inside of envelope; involved in assembly; interacts with viral RNPs and NS2 |
| M2 | 11,000 | 20–60 | Integral membrane protein; ion channel; essential for virus uncoating; from spliced mRNA | ||
| 8 | 890 | NS1 | 26,800 | 0 | Nonstructural; high abundance; inhibits pre-mRNA splicing; reduces interferon response |
| NS2 | 14,200 | 130–200 | Minor component of virions; nuclear export of viral RNPs; from spliced mRNA | ||
Influenza virus particles contain nine different structural proteins. The nucleoprotein (NP) associates with the viral RNA to form a ribonucleoprotein (RNP) structure 9 nm in diameter that assumes a helical configuration and forms the viral nucleocapsid. Three large proteins (PB1, PB2, and PA) are bound to the viral RNP and are responsible for RNA transcription and replication. The matrix (M1) protein, which forms a shell underneath the viral lipid envelope, is important in particle morphogenesis and is a major component of the virion (~40% of viral protein).
A lipid envelope derived from the cell surrounds the virus particle. Two virus-encoded glycoproteins, hemagglutinin (HA) and neuraminidase (NA), are inserted into the envelope and are exposed as spikes about 10 nm long on the surface of the particle. These two surface glycoproteins determine antigenic variation of influenza viruses and host immunity. The HA represents about 25% of viral protein and the NA about 5%. The M2 ion channel protein and the NS2 protein are also present in the envelope but at only a few copies per particle.
Because of the segmented nature of the genome, when a cell is coinfected by two different viruses of a given type, mixtures of parental gene segments may be assembled into progeny virions. This phenomenon, called genetic reassortment, may result in sudden changes in viral surface antigens—a property that explains the epidemiologic features of influenza and poses significant problems for vaccine development.
Influenza viruses are relatively hardy in vitro and may be stored at 0–4°C for weeks without loss of viability. Lipid solvents, protein denaturants, formaldehyde, and irradiation destroy infectivity. Both infectivity and hemagglutination are more resistant to inactivation at alkaline pH than at acid pH.
Genus Influenzavirus A contains human and animal strains of influenza type A, Influenzavirus B contains human strains of type B, and Influenzavirus C contains influenza type C viruses of humans and swine.
Antigenic differences exhibited by two of the internal structural proteins, the nucleocapsid (NP) and matrix (M) proteins, are used to divide influenza viruses into types A, B, and C. These proteins possess no cross-reactivity among the three types. Antigenic variations in the surface glycoproteins, HA and NA, are used to subtype type A viruses.
The standard nomenclature system for influenza virus isolates includes the following information: type, host of origin, geographic origin, strain number, and year of isolation. Antigenic descriptions of the HA and the NA are given in parentheses for type A. The host of origin is not indicated for human isolates, such as A/Hong Kong/03/68(H3N2), but it is indicated for others, such as A/swine/Iowa/15/30(H1N1).
So far, 18 subtypes of HA (H1–H18) and 11 subtypes of NA (N1–N11), in many different combinations, have been recovered from humans and animals.
The Orthomyxoviridae family also contains the genus Thogotovirus, members of which are not known to cause disease in humans.
The HA protein of influenza virus binds virus particles to susceptible cells and is the major antigen against which neutralizing (protective) antibodies are directed. Variability in HA is primarily responsible for the continual evolution of new strains and subsequent influenza epidemics. HA derives its name from its ability to agglutinate erythrocytes under certain conditions.
The primary sequence of HA contains 566 amino acids (Figure 39-2A). A short signal sequence at the amino terminal inserts the polypeptide into the endoplasmic reticulum; the signal is then removed. The HA protein is cleaved into two subunits, HA1 and HA2, that remain tightly associated by a disulfide bridge. A hydrophobic stretch near the carboxyl terminal of HA2 anchors the HA molecule in the membrane, with a short hydrophilic tail extending into the cytoplasm. Oligosaccharide residues are added at several sites.
FIGURE 39-2
Influenza virus hemagglutinin (HA) and neuraminidase (NA) surface glycoproteins. A: Primary structures of HA and NA polypeptides. The cleavage of HA into HA1 and HA2 is necessary for virus to be infectious. HA1 and HA2 remain linked by a disulfide bond (S–S). No posttranslational cleavage occurs with NA. Carbohydrate attachment sites • are shown. The hydrophobic amino acids that anchor the proteins in the viral membrane are located near the carboxyl terminal of HA and the amino terminal of NA. B: Folding of the HA1 and HA2 polypeptides in an HA monomer. Five major antigenic sites (sites A–E) that undergo change are shown as shaded areas. The amino terminal of HA2 provides fusion activity (fusion peptide). The fusion particle is buried in the molecule until it is exposed by a conformational change induced by a low pH. C: Structure of the HA trimer as it occurs on a virus particle or the surface of infected cells. Some of the sites involved in antigenic variation are shown (A). Carboxyl terminal residues (C) protrude through the membrane. D: Structure of the NA tetramer. Each NA molecule has an active site on its upper surface. The amino terminal region (N) of the polypeptides anchors the complex in the membrane. (Redrawn with permission from [A, B] Murphy BR, Webster RG: Influenza viruses, pp. 1185 and 1186, and [C, D] Kingsbury DW: Orthomyxo-and paramyxoviruses and their replication, pp. 1163 and 1172. In Fields BN [editor-in-chief] Virology. Raven Press, 1985.)
The three-dimensional structure of the HA protein has been revealed by x-ray crystallography. The HA molecule is folded into a complex structure (Figure 39-2B). Each linked HA1 and HA2 dimer forms an elongated stalk capped by a large globule. The base of the stalk anchors it in the membrane. Five antigenic sites on the HA molecule exhibit extensive mutations. These sites occur at regions exposed on the surface of the structure, are apparently not essential to the molecule’s stability, and are involved in viral neutralization. Other regions of the HA molecule are conserved in all isolates, presumably because they are necessary for the molecule to retain its structure and function.
The HA spike on the virus particle is a trimer composed of three intertwined HA1 and HA2 dimers (Figure 39-2C). The trimerization imparts greater stability to the spike than could be achieved by a monomer. The cellular receptor binding site (viral attachment site) is a pocket located at the top of each large globule. The pocket is inaccessible to antibody.
The cleavage that separates HA1 and HA2 is necessary for the virus particle to be infectious and is mediated by cellular proteases. Influenza viruses normally remain confined to the respiratory tract because the protease enzymes that cleave HA are expressed only at those sites. Examples have been noted of more virulent viruses that have adapted to use a more ubiquitous enzyme, such as plasmin, to cleave HA and promote widespread infection of cells. The amino terminal of HA2, generated by the cleavage event, is necessary for the viral envelope to fuse with the cell membrane, an essential step in the process of viral infection. Low pH triggers a conformational change that activates the fusion activity.
The antigenicity of NA, the other glycoprotein on the surface of influenza virus particles, is also important in determining the subtype of influenza virus isolates.
The spike on the virus particle is a tetramer composed of four identical monomers (Figure 39-2D). A slender stalk is topped with a box-shaped head. There is a catalytic site for NA on the top of each head, so that each NA spike contains four active sites.
The NA functions at the end of the viral replication cycle. It is a sialidase enzyme that removes sialic acid from glycoconjugates. It facilitates release of virus particles from infected cell surfaces during the budding process and helps prevent self-aggregation of virions by removing sialic acid residues from viral glycoproteins. It is possible that NA helps the virus negotiate through the mucin layer in the respiratory tract to reach the target epithelial cells.
Influenza viruses are remarkable because of the frequent antigenic changes that occur in HA and NA. Antigenic variants of influenza virus have a selective advantage over the parental virus in the presence of antibody directed against the original strain. This phenomenon is responsible for the unique epidemiologic features of influenza. Other respiratory tract agents do not display significant antigenic variation.
The two surface antigens of influenza undergo antigenic variation independent of each other. Minor antigenic changes are termed antigenic drift; major antigenic changes in HA or NA, called antigenic shift, result in the appearance of a new subtype (Figure 39-3). Antigenic shift is most likely to result in an epidemic.
FIGURE 39-3
Antigenic drift and antigenic shift account for antigenic changes in the two surface glycoproteins (hemagglutinin [HA] and neuraminidase [NA]) of influenza virus. Antigenic drift is a gradual change in antigenicity caused by point mutations that affect major antigenic sites on the glycoprotein. Antigenic shift is an abrupt change caused by genetic reassortment with an unrelated strain. Changes in HA and NA occur independently. Internal proteins of the virus, such as the nucleoprotein (NP), do not undergo antigenic changes.
Antigenic drift is caused by the accumulation of point mutations in the gene, resulting in amino acid changes in the protein. Sequence changes can alter antigenic sites on the molecule such that a virion can escape recognition by the host’s immune system. The immune system does not cause the antigenic variation but rather functions as a selection force that allows new antigenic variants to expand. A variant must sustain two or more mutations before a new, epidemiologically significant strain emerges.
Antigenic shift reflects drastic changes in the sequence of a viral surface protein, caused by genetic reassortment between human, swine, and avian influenza viruses. Influenza B and C viruses do not exhibit antigenic shift because few related viruses exist in animals.
The replication cycle of influenza virus is summarized in Figure 39-4. The viral multiplication cycle proceeds rapidly. There is the shut-off of host cell protein synthesis by about 3 hours postinfection, permitting selective translation of viral mRNAs. New progeny viruses are produced within 8–10 hours.