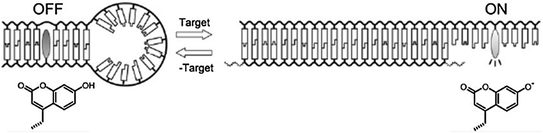
Fig. 27.1
a Molecular structure of a pyrene-functionalized cationic oligopeptide. b Binding of the beacon to double-stranded DNA. c Schematic working mechanism (Recreated with kind permission from American Chemical Society; J. Am. Chem. Soc. 2012; 134(4):1958–61)
27.1.2 Photodynamic Molecular Beacons
With the same sensitive detection ability as molecular beacons, photodynamic molecular beacons feature light-activated singlet oxygen generating properties of photosensitizers, which are altered, reduced, or intensified by molecular recognition or environmental changes. Historically, development of photodynamic molecular beacons has not been based on nucleic acids. For instance, activatable photosensitizers can control how singlet oxygen is quenched and activated via protease control [21]. Cytotoxic singlet oxygen is only produced upon irradiation if the photosensitizer is liberated from the quencher. Compared to fluorophores of conventional molecular beacons, the excited singlet state can alternatively enter a lower-energy triplet state via intersystem crossing [12]. Surrounded by diffused oxygen molecules, triplet-state photosensitizers can interact with these to yield singlet oxygen [22]. As for the photosensitizer spectral properties, those absorbing at near-infrared wavelengths (650–900 nm) are usually selected since deeper tissue penetration and lower background fluorescence are possible in this range due to intrinsic optical properties of tissues [23]. The localization of photosensitizers is also important, and partitioning into organelles like the mitochondria and endoplasmic reticulum can improve efficacy [22, 24]. The principles in designing photodynamic molecular beacons for controlling singlet oxygen generation are essentially the same as for fluorescence beacons and must enable beacon activation upon target recognition [25, 26].
DNA-based activatable photosensitizers have been constructed as a means to control on-and-off switching of singlet oxygen in response to nucleic acid target binding [27]. Pyropheophorbide and Black Hole Quencher were conjugated to two separate complimentary oligonucleotide strands to serve as photosensitizer and quencher, respectively. Both fluorescence emission and singlet oxygen production were almost completely shut down, while an excess of complimentary sequence was able to recover 85 % of the singlet oxygen after hybridization. Figure 27.2 shows the conversion of a classic molecular beacon into a photodynamic one [28]. Further innovations include a pH-sensitive DNA i-motif controlling photosensitized singlet oxygen [29]. The i-motif quadruplex was able to keep the photosensitizer and quencher in close proximity in the absence of acidic conditions. The formation of the triplet state of the sensitizer was prevented by quenching. The i-motif was not stable when the pH was raised above five, resulting in increased distance between photosensitizer and quencher and increased singlet oxygen production. This approach successfully permits the modulation of photodynamic activity in response to pH change. A peptide photodynamic molecular beacon was synthesized for the diagnosis and treatment of epithelial cancer [30]. The photocytotoxicity is triggered via fibroblast activation protein (FAP), which is a cell-surface serine protease in cancer-related fibroblasts of human epithelial carcinomas but not healthy tissues. The peptide loop specific to FAP could be cleaved and allowed restoration of fluorescence and singlet oxygen. Specific photocytotoxicity was observed toward HEK cells expressing FAP, demonstrating a feasible therapeutic option as well.
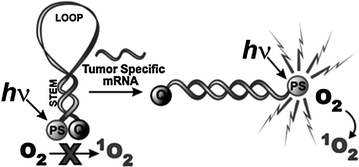

Fig. 27.2
The concept of mRNA photodynamic molecular beacon (Recreated with kind permission from Royal Society of Chemistry; Photochem. Photobiol. Sci. 2008; 7(7):775–81)
As photosensitizers are also fluorescent, they can be used for imaging combined with photodynamic therapy (PDT). PDT is a minimally invasive therapeutic method that avoids repeated dosage resistance and other side effects [22, 24]. PDT aims at destroying target cells using three elements: light irradiation, photosensitizers, and cytotoxic singlet oxygen [31]. Reactive oxygen species activated from irradiated photosensitizers are responsible for killing targeted cells. PDT requires careful planning to limit damage to surrounding healthy tissues. PDT and imaging can be conveniently carried out with a single fluorescent photosensitizer, with porphyrins being a good example [32]. Porphyrins have a long history as tumor-specific diagnostic fluorescence imaging agents. For PDT applications, porphyrins meet the demands of good availability, conjugation compatibility, quencher compatibility, singlet oxygen quantum yield, and fluorescence quantum yield [22]. Thus, for photodynamic beacons, porphyrin-related compounds are suitable candidates to use in engineer beacons. Quenchers are important for photodynamic molecular beacons since any unquenched beacons might exert unwanted singlet oxygen on healthy tissues under irradiation. Thus, the signal-to-noise ratio is important for therapeutic applications. Carotenoid (CAR) has been studied as a unique quencher since it is able to directly scavenge singlet oxygen [33]. CAR has dual quenching ability, which helps to improve beacon efficacy; photosensitizer excited-state deactivation and singlet oxygen scavenging.
For photodynamic beacons, a linker sequence specific to the target, such as a tumor-specific enzyme, can act as the recognition probe. The goal is to trigger the enzymatic cleavage of the linker in disease-specific cells to restore light-induced singlet oxygen, which is quenched in the inactive beacon. Figure 27.3 illustrates an engineered photodynamic beacon targeting breast cancer cells [34]. Breast cancer is a common disease and requires more effective diagnosis and treatment options. The proposed photodynamic peptide-based beacon offers promise for non-invasive and site-selective treatment. In this approach, the beacon was able to be activated by matrix metalloproteinases (MMPs), proteases implicated in the disease. Another example was a membrane-permeable, cancer-specific beacon with a built-in apoptosis sensor to carry out therapeutic and imaging functions [35]. The beacon contained a fluorescent pyromoiety as a photosensitizer and a BHQ-3 quencher connected by the caspase-3-cleavable sequence (KGDEVDGSGK), while folate served as a targeting moiety. The folate-carrying beacon preferentially accumulated where folate receptors were overexpressed. Following uptake, the next step involved light activation and singlet oxygen was generated. Apoptosis led to the activation of caspase-3, which specifically cleaved the beacon. Thus, dying cells were able to be identified by the restoration of photosensitizer emission. Although measurements of singlet oxygen luminescence and photobleaching of photosensitizers have been taken to gauge tissue damage during the course of PDT treatment, these methods only behave indirectly to give feedback. A subsequent approach made use of 5-carboxy-X-rhodamine (Rox) as a donor, along with the acceptor pyro and a caspase-3-specific peptide sequence. This beacon was more effectively able to both induce and detect apoptosis [36]. This represents a new strategy that enables light to be regulated to activate photosensitizer dosage with direct molecular feedback. Another approach involved the combination of photosensitizers with single-walled carbon nanotubes as quenchers [37]. Single-walled carbon nanotubes have been regarded as efficient quenchers [38] and intracellular carriers [39]. Chlorine 6 was attached to a single-stranded aptamer for fluorescence and singlet oxygen generation. Upon binding to single-walled carbon nanotubes, fluorescence and light-activated singlet oxygen were quenched. Targets that bound to the aptamer modulated its interaction with single-walled carbon nanotubes and released the Ce6 from quenched state. Irradiation then caused the production of singlet oxygen.
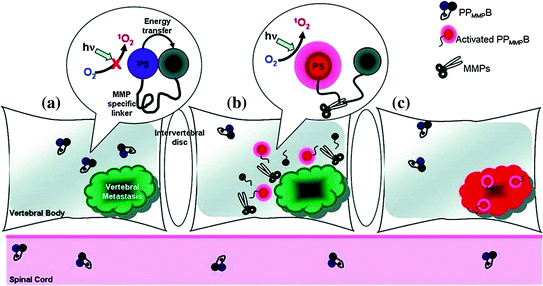

Fig. 27.3
Three phases of molecular beacon triggered by vertebral metastases. a Molecular beacons are quenched when accumulated in tissue. b MMPs within vertebral metastases cleave the loop portion of the beacon. c Beacons relieved from quenched status restore fluorescence and trigger singlet oxygen (Recreated with kind permission from American Chemical Society; Bioconjugate Chem. 2011; 22(6):1021–30)
The strategy of singlet oxygen control with photodynamic molecular beacons is no longer limited to classic designs. An X-ray-activated system that used a lanthanum fluoride nanoparticle covalently linked to a photosensitizer was capable of yielding singlet oxygen. This approach offers opportunity for the application of photodynamic beacons deep in human tissue [40].
27.1.3 Thermodynamic Properties of Molecular Beacons
Most studies of molecular beacon thermodynamics have been related to nucleic acid-based beacons. Figure 27.2 demonstrates the first nucleic acid-based photodynamic molecular beacon [28], which follows the same principles as standard molecular beacons. To investigate thermodynamic effects of molecular beacons, an understanding of a three-state transition model of classic molecular beacons, based on temperature, is useful to avoid false positives (activation in the absence of targets) and false negatives (no activation with targets present) [8]. The thermal denaturation profile of molecular beacons without targets is shown in Fig. 27.4. Within the low-temperature phase, molecular beacons exist as loop–stem structures. Gradual increase in temperature results in the transition from a hairpin-loop structure to a random-coil structure, as indicated as Phase 3. Note that in this case, the increase in fluorescence with temperature increase is not due to hybridization. High fluorescence can be achieved when all molecular beacons come to this completely unquenched state. Hybridization can take place between molecular beacons and target oligonucleotides at temperatures below the DNA hybrid melting temperature (Phase 1). The high fluorescence of the hybridized beacons (compared to that of closed molecular beacons at corresponding temperatures) drops noticeably with ascending temperature, due to breaking of hydrogen bonds that form hybridized double helix. The dehybridization of the target oligonucleotides enables the remaining single-stranded molecular beacons to recover to stable stem–loop structures with fluorophores becoming quenched again. This fluorescence turning point indicates the transition of molecular beacons back into hairpin structures (Phase 2). A portion of molecular beacons does not undergo transition from Phase 1 to Phase 2 at all and become randomly coiled, that is, going from Phase 1 directly to Phase 3. Eventually, all molecular beacons tend to open to a random-coiled phase as the temperature is elevated. It is interesting that fluorescence of random-coiled molecular beacons (Phase 3) is lower than that of hybridization status (Phase 1) and can be attributed to incomplete quenching of fluorophores in the random-coil form. Molecular beacons with a single-nucleotide mismatch target experience similar transitions but with a lower transition temperature at the turning point. This permits discrimination between matched and mismatched targets and demonstrates the specificity and selectivity of molecular beacons [8].
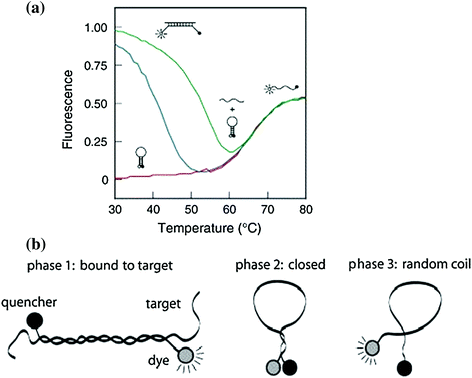

Fig. 27.4
Phase transition of molecular beacons in solutions. a Molecular beacons denaturation curves: solo molecular beacons without target nucleotides (red curve), with perfectly complementary targets (green curve) and with excess mismatched targets (blue curve) (Recreated with kind permission from Elsevier; real-time assays with molecular beacons and other fluorescent nucleic acid hybridization probes. Clin. Chim. Acta. 2006 Jan; 363(1–2):48–60. b Schematic representation of three transition states. Phase 1: hybridization hybrid between molecular beacons and targets. Phase 2: independent molecular beacons without targets. Phase 3: molecular beacons melt into random-coiled state. Recreated with kind permission from Oxford University Press; Nucl. Acids Res. 2003; 15,31(4):1319–30)
Melting temperature (Tm), the most important property in beacon transitions, is defined as the temperature at which 50 % of the strands are hybridized in the double-helix state [41]. Numerous factors are responsible for determining Tm: base stacking [42], guanine–cytosine content (GC content), buffer composition, stem length, and secondary structure [2, 8]. Since the thermodynamic stability of the G-C bond is greater than the A-T bond, rich GC content sequences tend to have a higher Tm [41, 43, 44]. A longer stem length is found to change the melting profile by broadening the transition between free molecular beacons in loop–stem structure to hybridized double helices [9]. Interactions between nucleotides within the same strand may occur with secondary internal loops. These loops are regarded as competitors against target oligonucleotide binding and decrease the Tm of the hybrid. General thermodynamic analysis of ion effects has been studied in terms of hairpin structure, indicating that reduced ion concentration may add to the loop free energy and destabilize the structure [45], but once the stabilization effect is saturated, a higher ion concentration does not further enhance stability [46]. As to the effect of target probe length, Tm generally grows with longer probe lengths. Stem length also exerts significant influence on the Tm. A 2.7 °C reduction in hybrid Tm was observed by increasing the stem length by one base [9]. Thermodynamic analysis of molecular beacons is important due to the fact that it is closely related to the selectivity of molecular beacons [11]; the difference between phase transition temperature of a perfect beacon–target double helix and a mismatched duplex determines the selectivity of any given molecular beacon.
27.2 Classic Molecular Beacon Design Considerations
Thanks to vast online bioinformatics tools and database resources such as basic local alignment search tool (BLAST), we are able to target various wild-type gene sequences from huge databases and use the corresponding complementary sequences as beacon probes. However, several considerations should be taken to carefully design a molecular beacon. In this section, some of these general design considerations of classic molecular beacons will be discussed. Recent progress in molecular beacons involves new ideas in beacon design either to enhance their performance or to expand their applications.
The probe length, typically a 15–25-base oligonucleotide [47], should be of suitable length in order to position the fluorophore as far as possible from the quencher upon hybridization, or to be at least twice the length of each stem sequence [1]. Longer probes are also beneficial for faster hybridization kinetics. However, the different requirement of GC content between probe and stem portions is a barrier that makes this challenging. Bounded by three hydrogen bonds, GC pairs are more stable than AT pairs, which have only two hydrogen bonds. GC pairs should comprise at least half of the stem pairs in order to form a secure, rigid hairpin structure, while too much overall GC content should be avoided to avoid secondary internal loops formation. These loops, however, may appear even though GC pairs are not dominating, for example, consecutive AT pairs may also break the loop into secondary ones. To minimize the sliding of the hybridized target along the probe, GC residues are expected to occupy end positions of the loop [8]. Given that a wild-type sequence is a natural genetic code, it is not always straightforward to select sequences meeting all requirements, although the relative long length of genes compared to the molecular beacon permits many options. The stem portion is less difficult to design, with respect to length and GC content. Longer stems contribute to provide discrimination between mutant mismatch targets and perfectly matched ones within a large temperature range and increase the Tm gap between perfectly matched duplexes and mismatched ones. However, longer stems require longer loop sequences to overcome the energy barrier encountered in unzipping the stems and a decrease in hybridization rate may be observed [8]. They may also lead to false-negative results, whereas short and unstable stems give false-positive signals and higher background fluorescence [2, 9].
Fluorophores, analogous photosensitizers, and their quenchers should be selected carefully. Overall, fluorophores and photosensitizers should be chosen to have maximum emission spectral overlap with quencher absorption. Good quenching efficiency is required so that accuracy and sensitivity in fluorescence detection can be achieved with a low background signal. Fortunately, many quenchers like dabcyl can be applied to a very wide range of fluorophores. These universal quenchers usually exert both FRET and contact quenching [47]. Selection of fluorophores is based on various criteria [48]: an easily tuned photophysical and spectral range, stability against chemical, thermal, or light effects, and good bioconjugation properties. To avoid being quenched by nucleotides, it is best not to link fluorophores next to guanosine bases, which themselves can act like quenchers [10, 49].
Apart from design strategies, specific experimental conditions are important to consider. For example, the reaction buffer can have profound effects on performance. Since nucleic acid backbones are strongly negatively charged, cations such as sodium and magnesium help balance the charge so that molecular beacons are properly folded and florescence is quenched before the addition of target oligonucleotides. Ion concentration and size are important factors [46, 50]. For in vitro analysis, most molecular beacons are used in aqueous solutions. Organic solvents also have been studied as a buffer for molecular beacons. Although hybridization reactions in most common organic solvents do not take place to a full extent, faster kinetics are observed. Tm is found to have a solvent concentration-dependent decrease [51]. Fortunately, there are several online simulation tools that can assist the design of DNA molecular beacons, for example, the UNAFold Web Server of University at Albany [52]. These tools are able to give predicted thermodynamic properties and schematic hybridization possibilities, which can greatly lower the potential of unwanted secondary structure during the design process.
27.3 Next-Generation Molecular Beacon Designs
Numerous publications regarding optimization of molecular beacons have described enhancements to the performance of molecular beacons and have largely expanded the types of applications they are used in. In this section, we will introduce some of the more recent developments in design, from adjusting the beacon itself to incorporation with other technologies. These efforts not only improve molecular beacon efficacy but also reveal innovative ideas that may serve as future engineering options for photodynamic molecular beacons.
27.3.1 Optimization of Fluorophores and Quenchers
Since activatable photosensitizers share similar mechanisms with normal fluorophores of molecular beacons, approaches for selecting photosensitizers can benefit from examining recent trends in the selection of fluorophores and quenchers for standard molecular beacons [22]. Compared to the challenge of designing an optimal beacon stem and loop sequence, it is more straightforward to optimize fluorophores and quenchers; for example, by using fluorophores with more desirable spectral properties or by using quenchers that have higher quenching efficiency. As mentioned above, the efficacy of certain quenchers can be predicted by spectral properties alone. To further enhance sensitivity of molecular beacons, a “super-quencher” array of multiple quenchers was proposed [53]. 99.7 % quenching efficiency was observed using super-quenchers, which may result from higher absorption efficiency and the increased dipole–dipole coupling between the quenchers and the fluorophores. The architecture of super-quenchers can be modified as multiple quenchers linked to a trebler phosphoramidite that branches one end of the beacon stem or a series of quenchers linearly connected to a photodynamic beacon [54]. Further development of this beacon led to programmed lipoprotein and liposome nanoparticle aggregation due to the hydrophobic character of the super-quenchers, which is inserted into the nanoparticles to induce irreversible aggregation upon target nucleic acid recognition [55].
Other than changes in type and number of quenchers, improvements can be achieved with other design aspects. Synthesis of classic molecular beacons involves two steps: conjugation of quenchers and conjugation of fluorophores [1]. However, a deprotection step is often required between these two steps, and therefore, more cost-effective and simplified conjugation schemes are useful [56]. Quencher-free molecular beacons simplify beacon design and synthesis [57]. The first example of this approach came from attaching a fluorophore to the loop portion so the probe provided a fluorescence differential to discriminate between perfectly matched targets and single-base mismatches [58]. Other quencher-free molecular beacons were subsequently reported. Analogous to contact quenching of conventional molecular beacons, formation of H-type dimers by two identical dye molecules tethered to both ends allowed silencing of fluorescence with restoration upon hybridization [59]. Similarly, a spectral shift from excimer fluorescence to monomer fluorescence with a different emission wavelength is feasible [60, 61]. These so-called excimer–monomer switching (EMS) molecular beacons differ from non-fluorescent dye dimerization beacons since the fluorescence intensity ratio between excimer and monomer emissions becomes the key discriminating criteria instead of total fluorescence intensity. Fluorescent groups themselves are able to act as “quenchers”, with terminal fluorophores serving as quenchers for each other. The determination of beacon opening is based on the ratio of fluorescence intensity emitted by the two fluorophores at their respective wavelengths [62]. This type of molecular beacon functions via wavelength shifting. Molecular beacons that do not have typical quenchers exist. As previously mentioned, certain nucleotide bases, especially guanosine, can quench fluorescence, so it is possible that nucleotide bases can take the role of quenchers in well-designed schemes. This is because these nucleotide bases tend to transfer electrons to fluorophores in terms of a photoelectron transfer [63]. The geometry of the attached fluorophores is important to take full advantage of nucleotide bases as quenchers. An interesting design used a fluorophore tethered to the center of the loop, accompanied by two guanosines on either side, to generate effective quenching [64]. Development of a quencher-free molecular beacon that has a fluorophore within its stem portion was also reported [56]. In this case, the microenvironment plays an important role in quenching. Making use of the fact that some fluorophores have solvent polarity preferences and there is a hydrophobic environment within the hybridization duplex between beacons and targets, fluorescence can be switched on or off based on the position of fluorophores. A linear probe with such strategy was synthesized [65]. As shown in Fig. 27.5, this approach was carried out so that hybridization controlled the local environment of the dye molecule, which was attached to the middle of one stem [56]. 7-hydroxycoumarin, whose fluorescence is controlled by protonation or deprotonation, was used as the fluorophore. A similar approach made use of thiazole orange (TO) and thiazole red (TR) adjacently embedded on two arms of the stem. The incorporation of two fluorophores makes the beacon emit red when the beacon is closed and the fluorophores are kept within the hydrophobic stem. When the complementary target binds, the stem opens and shifts the red emission to a blue one. This “DNA Traffic Light” approach successfully combined concepts of wavelength shifting and in-stem labeling together to produce a smart probe that is suitable for living cell applications. These combined strategies help to reduce false-positive and false-negative signals given from autofluorescence or undesired quenching, which usually occur in single-emission molecular beacons [66]. Photodynamic molecular beacons could take advantage of the same approaches.