CHAPTER 36 Motor Proteins
Molecular motors use ATP hydrolysis to power movements of subcellular components, such as organelles and chromosomes, along the two polarized cytoskeletal fibers: actin filaments and microtubules. No motors are known to move on the apolar intermediate filaments. Motor proteins also produce force locally within the network of cytoskeletal polymers, which transmits these forces to determine the shape of each cell and, ultimately, the architecture of tissues and whole organisms. Chapters 37 to 39 and 44 illustrate how motors move cells and their parts.
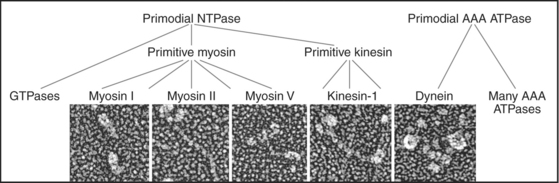
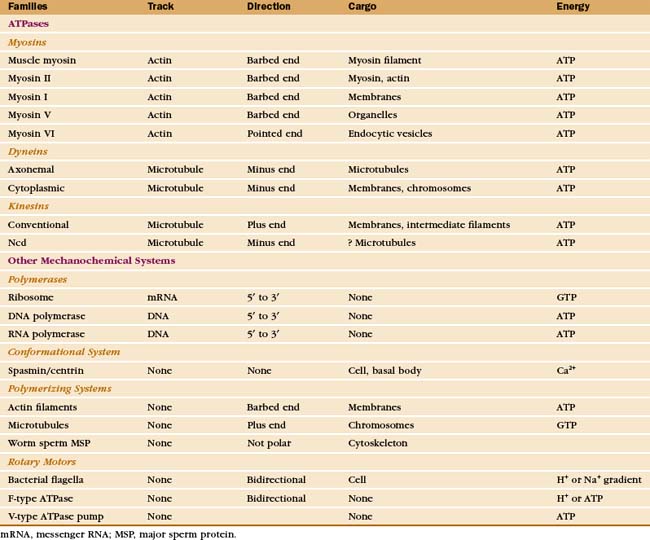
Just three families of motor proteins—myosin, kinesin, and dynein—power most eukaryotic cellular movements (Fig. 36-1 and Table 36-1). During evolution myosin, kinesin, and Ras family GTPases appear to have shared a common ancestor (Fig. 36-1), whereas dynein is a member of the AAA ATPase family (Box 36-1). Although the ancestral genes appeared in prokaryotes, and prokaryotes have homologs of both actin and tubulin, none of these motor proteins has been found in prokaryotes. Over time, gene duplication and divergence in eukaryotes gave rise to multiple genes for myosin, dynein, and kinesin, each encoding proteins with specialized functions. Even the slimmed down genome of budding yeast includes genes for five myosins, six kinesins, and one dynein. Table 36-1 lists other protein machines that produce molecular movements during protein and nucleic acid synthesis, proton pumping, and bacterial motility.
BOX 36-1 AAA ATPases
The common ancestor of life on the earth had a gene for a versatile ATP-binding domain. Through gene duplication and divergence this progenitor gave rise to the AAA family of ATPases in all branches of the phylogenic tree. Given the remarkable variety of functions of the contemporary proteins, the name “ATPases Associated with Diverse Activities” is apt. The family now includes regulatory subunits of proteasomes (see Fig. 23-5); proteases from prokaryotes, chloroplasts, and mitochondria; Hsp100 protein folding chaperones; dynein microtubule motors (Fig. 36-14); the microtubule severing protein katanin (see Fig. 34-9); activators of origins of replication (including ORC1, 4, and 5 and Mcm-7 [see Fig. 42-7]); clamp loader proteins for DNA polymerase processivity factors (see Fig. 42-11); two proteins required for peroxisome biogenesis (see Appendix 18-1); and proteins involved in vesicular traffic such as NSF (the N-ethylmaleimide-sensitive factor [see Fig. 21-12]).
Motor proteins have two parts: a motor domain that utilizes adenosine triphosphate (ATP) hydrolysis to produce movements and a tail that allows the motors to self-associate or to bind particular cargo. Within the three families, the tails are more diverse than the motor domains, allowing for specialized functions of each motor isoform.
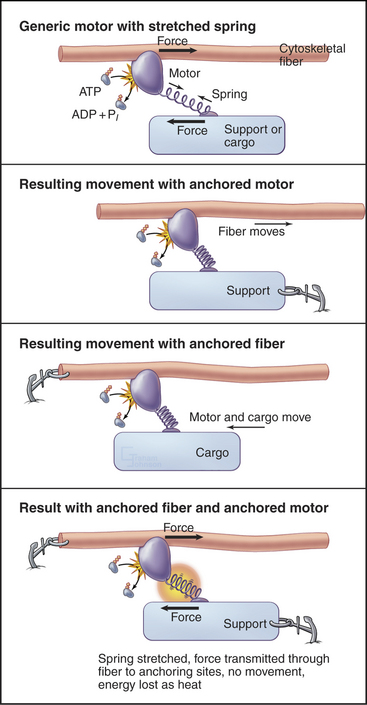
All motor proteins are enzymes that convert chemical energy stored in ATP into molecular motion to produce force upon an associated cytoskeletal polymer (Fig. 36-2). If the motor is anchored, the polymer may move. If the polymer is anchored, the motor and any attached cargo may move. If both are anchored, the force stretches elastic elements in the molecules transiently, but nothing moves, and the energy is lost as heat. Cells use all of these options (see Chapters 37to3839).
Biochemists originally discovered and purified these motors by means of enzyme activity or in vitro motility assays (Fig. 36-11). With the prototype enzymes identified, investigators found further examples and variant isoforms of each motor by purification of the proteins, molecular cloning of DNAs, genomewide DNA sequencing, or genetic screening.
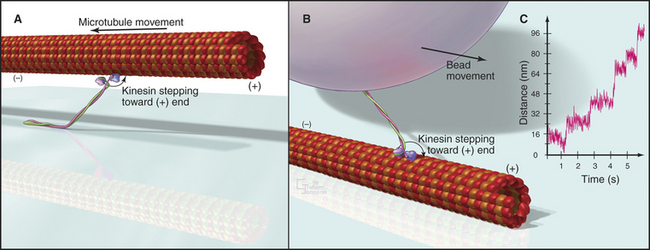
Figure 36-11 in vitro motility assays for microtubule motors. A, Gliding assay. Kinesin or dynein that is attached to a microscope slide uses ATP hydrolysis to move microtubules over the surface. A single kinesin-1 molecule can move a microtubule in this assay. Microtubules can be imaged by video-enhanced differential interference contrast microscopy (see Fig. 34-7).B, Bead assay. Kinesin or dynein that is attached to a plastic bead uses ATP hydrolysis to move the bead along a microtubule attached to the microscopic slide. C, Experimental measurement of the kinesin-1 step size using the bead assay. The bead is held in a laser optical trap so that 8-nm steps can be recorded, as a single, two-headed kinesin-1 moves a bead processively along a microtubule, as in B. The position of the bead is recorded with nanometer precision by interferometry.
(Reference: Svoboda K, Schmidt CF, Schnapp BJ, Block SM: Direct observation of kinesin stepping by optical trapping interferometry. Nature 365:721–727, 1993.)
Myosins
Myosins are the only motors that are known to use actin filaments as tracks. As is discussed in the section entitled “The Myosin Superfamily,” the members of this diverse family arose from a common ancestor and share a common motor unit called a myosin “head” that produces force on actin filaments (Fig. 36-3). Myosins have one or two heads attached to various types of tails that are adapted for diverse purposes, including polymerization into filaments, binding membranes, and interacting with various cargos.
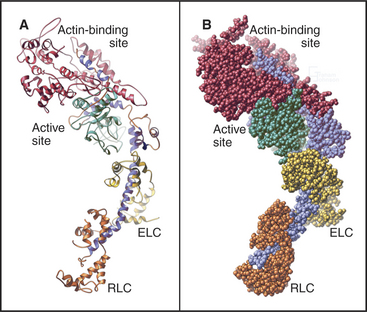
Myosin heads consist of two parts. A catalytic domain at the N-terminus of the myosin heavy chain binds and hydrolyzes ATP and interacts with actin filaments. Light chain domains consist of an α-helical extension of the heavy chain from the catalytic domain associated with one to seven light chains. Calmodulin (see Fig. 3-12) serves as a light chain for some cytoplasmic myosins, but many light chains are specialized relatives of calmodulin.
Myosin Mechanochemistry
Studies of skeletal muscle myosin established general principles that apply, with some interesting variations, to energy transduction by all myosins. This founding member of the myosin family is responsible for the forceful contraction of skeletal muscle. Like other types of myosin-II, it has two heads on a long tail formed from an α-helical coiled-coil. These tails polymerize into bipolar filaments (see Figs. 5-7 and 39-6).
The head of muscle myosin was originally isolated as a proteolytic fragment called subfragment-1 (Fig. 36-3). The N-terminal 710 residues of the heavy chain form the globular catalytic domain. The nucleotide binding site in the core of the catalytic domain is formed by a β-sheet flanked by α-helices with a topology similar to Ras GTPases (see Fig. 4-6) despite little sequence similarity. The γ-phosphate of ATP inserts deeply into the nucleotide-binding site with the adenine exposed on the surface. Actin binds more than 4 nm away from the nucleotide on the other side of the head. The light-chain domain has an essential light chain and a regulatory light chain wrapped around and stabilizing a long α-helix formed by the heavy chain (Fig. 36-3). The interaction of light chains with the heavy chain α-helix is similar to calmodulin binding its target proteins (see Fig. 3-12).
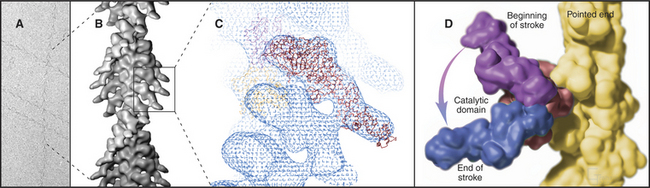
Myosin heads bind tightly and rigidly to actin filaments in the absence of ATP. This is called a rigor complex because it forms in muscle during rigor mortis when ATP is depleted after death. Myosin heads bound along an actin filament form a polarized structure, resembling a series of arrowheads when viewed from the side (Fig. 36-4). The heads bind at an angle and wrap around the filament. Their orientation defines the barbed and pointed ends of the actin filament (see Fig. 33-8). All known myosins except myosin-VI move toward the barbed end of the filament.
The atomic structures of the myosin head and actin filament fit nicely into the three-dimensional structure of the decorated filament determined by electron microscopy, providing a reasonable model of the complex at near atomic resolution (Fig. 36-4). The model shows each head in contact with two adjacent actins but does not reveal important details, including atomic contacts, between the proteins. This model is the structural starting point for understanding the mechanics of force production.
Actomyosin ATPase Cycle
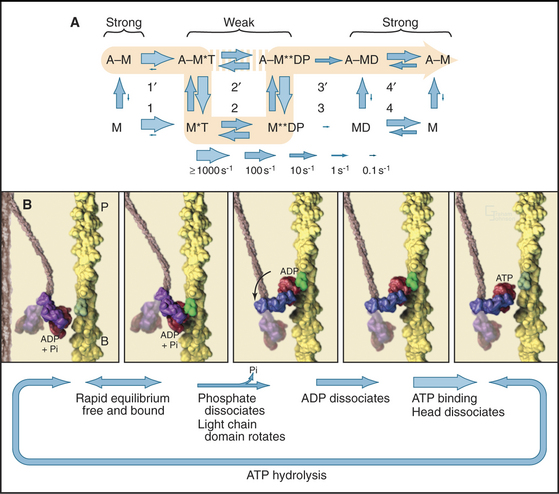
Myosin uses energy from ATP hydrolysis to move actin filaments, so an appreciation of the mechanism requires an understanding of the steps in the biochemical reaction. Figure 36-5A looks intimidating, but working through it one step at a time reveals its logic and simplicity. Note that the mechanism consists of two parallel lines of chemical intermediates. This series of reactions explains why myosin alone turns over ATP remarkably slowly, at a rate of only about 0.02 s−1. First, consider the bottom line, which shows how myosin hydrolyzes ATP in the absence of actin:
Now focus on the upper line in Figure 36-5A, where myosin is associated with an actin filament. The chemical intermediates are the same, but some of the key rate constants differ for the actin-bound and free enzymes. Steps 1 and 2 are similar to those of free myosin, but step 3—the dissociation of phosphate—is much faster when a head is bound to an actin filament. As a result, myosin bound to actin traverses the ATPase cycle about 200 times faster than myosin free in solution, and ATP hydrolysis becomes the rate-limiting step. This effect of actin is referred to as “actin activation of the myosin ATPase.” A practical advantage of this mechanism is that the ATPase cycle is essentially turned off unless a head interacts with an actin filament.
Finally, consider the vertical arrows representing transitions between bound and free states of each myosin chemical intermediate. All myosin intermediates bind rapidly to actin filaments, but the dissociation rate constants vary over a wide range depending on the nucleotide that is bound to the active site of the myosin. Myosin with no nucleotide or with bound ADP alone dissociates very slowly and therefore binds tightly to actin filaments. Myosin with bound ATP or ADP+Pi dissociates rapidly from actin, so these states bind actin weakly.
These rapid binding and dissociation reactions allow myosin intermediates (MT and MDP) to hop on and off actin filaments on a millisecond time scale, a key feature of muscle contraction (see Chapter 39). As a result of this rapid equilibrium, a single pathway cannot be drawn through the reaction mechanism of ATP, myosin, and actin. One cycle of ATP hydrolysis takes about 50 ms. Starting with AM, ATP binds very rapidly and sets up a rapid, four-way equilibrium including AMT, MT, AMDP, and MDP—the major intermediates during steady-state ATP turnover. Because the products of ATP hydrolysis dissociate much more rapidly from AMDP than from MDP, the favored pathway out of this four-way equilibrium is through AMDP to AMD and back to AM. Because the fraction of myosin heads bound to actin in the AMDP state depends on the actin concentration, the overall ATPase rate depends on the actin concentration. At the high actin concentrations in cells, a significant fraction of myosin heads are associated with actin (about 10% in contracting muscle), but each molecule continues to exchange on and off actin filaments.