Mechanisms of Nutrient Sensing1
Douglas G. Burrin
Teresa A. Davis
1Abbreviations: AMP, adenosine monophosphate; AMPK, adenosine monophosphate-activated protein kinase; ATF4, activating transcription factor 4; ATP, adenosine triphosphate; CCK, cholecystokinin; ChREBP, carbohydrate-response element-binding protein; 4EBP1, eukaryotic initiation factor 4E-binding protein 1; EE, enteroendocrine; eEF2, eukaryotic elongation factor 2; eIF, eukaryotic initiation factor; GβL, G-protein β-like protein; GCN2, general control non-derepressible 2; GI, gastrointestinal; GLP, glucagon-like peptides; GPCR, G-protein-coupled receptor; IGF, insulinlike growth factors; IRS-1/2, insulin receptor substrate-1/2; LC-PUFA, long-chain polyunsaturated fatty acid; LXR, liver X receptor; met-tRNAi, initiator methionyl-transfer RNA; mTOR, mammalian target of rapamycin; mTORC, mammalian target of rapamycin complex; MuRF1, muscle-specific ring finger protein 1; PGC-1α, peroxisome proliferator-activated receptor-γ coactivator-1α; PI3K, phosphoinositide 3-kinase; PKB, protein kinase B; PPAR, peroxisome proliferator-activated receptor; PPRE, peroxisome proliferator response element; PepT1, peptide transporter 1; S6K1, 70-kDa ribosomal protein S6 kinase; SGLT1, sodium/glucose cotransporter 1; SIRT1, silent information regulator T1; SNAT2, system A sodiumdependent neutral amino acid transporter 2; TSC1/2, tuberous sclerosis complex 1/2; X5P, xylulose 5-phosphate.
The term nutrient sensing has emerged to describe the molecular mechanisms by which nutrients and their metabolites interact with various cell surface receptors, intracellular signaling proteins, and nuclear receptors and modulate the activity of a complex network of signaling pathways that regulate the growth and function of cells. Nutrients also trigger the release of hormones and neurotransmitters that act on neighboring or distant cells through paracrine or endocrine mechanisms to regulate cellular growth and function. This chapter discusses some of the key nutrient-sensing mechanisms in different tissues within the body.
NUTRIENT SENSING BY THE GUT
Nutrient sensing in humans, as in most mammals, begins in the gastrointestinal (GI) tract, including the oral cavity. Nutrient sensing begins with taste buds that line the tongue and palate epithelium. This vital sensory process of tasting foods functions to recognize foods that are nutritious, giving a sweet taste, as well as ingested compounds that may be poisonous or harmful, giving a bitter taste (see also the chapter on nutrition and the chemical senses). Specialized cells and receptors present on taste buds mediate the five general taste sensations of sweet, sour, salty, bitter, and savory (also called umami) (1, 2). These taste sensations are produced within the taste buds by specialized taste receptor cells that express numerous surface receptors that recognize different chemicals.
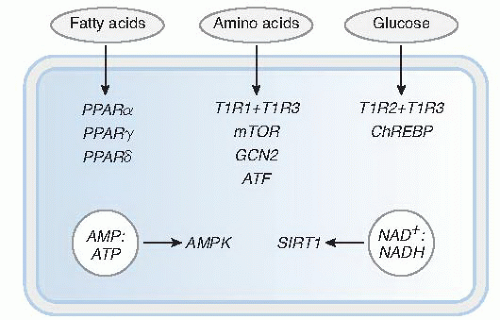
The taste sensations of sweet and umami are mediated by a family of three G-protein-coupled receptors (GPCRs) that function as taste receptors, namely, T1R1, T1R2, and T1R3, which form homodimeric or heterodimeric receptor complexes. Taste cells expressing the combination of T1R2 + T1R3 recognize sugars, artificial sweeteners, and some D-amino acids, and the result is a sweet taste (Fig. 47.1). Taste cells expressing the heterodimeric T1R1 + T1R3 receptor recognize the D-amino acids, glutamate and aspartate, and the result is the savory or umami taste. Other metabotropic receptors are found in taste cells and may mediate some of the umami taste (3, 4, 5). The bitter taste sensation is mediated by specific taste cells that express another family of GPCRs, namely T2R, that recognizes an array of bitter tastants, such as denatonium and quinine. Studies showed that the salty taste sensation is mediated by the epithelial sodium channel expressed on taste cells (6). Finally, the sour taste sensation is mediated by a member of the transient receptor potential ion channel family, PKD2L1, that functions as an acid receptor in taste cells.
Nutrient sensing beyond the oral cavity of the GI tract is accomplished by specialized, differentiated epithelial cells that sense the presence of nutrients in the stomach and intestinal lumen as they are processed after meal ingestion. These cells, called enteroendocrine (EE) cells, are one of four different cell lineages derived from stem cells that reside in the inner layer of the mucosal lining referred to as the crypt (7). The EE cells function as key
nutrient sensors within the mucosal wall that recognize carbohydrates, triglycerides, and protein in the gut lumen. The EE cells function to coordinate the recognition of luminal nutrients with activation of gut physiologic functions, such as motility, fluid secretion, and blood flow, by secreting hormones and neurotransmitters. More than 20 different EE cell types are recognized, and they differ in their gut location and type of hormones secreted (8). The EE cells differ from most other epithelial cells, such as parietal cell or enterocytes, that are programmed to produce acid and digestive enzymes to digest food components into their simplest, constituent units, namely, sugars, fatty acids, and amino acids.
nutrient sensors within the mucosal wall that recognize carbohydrates, triglycerides, and protein in the gut lumen. The EE cells function to coordinate the recognition of luminal nutrients with activation of gut physiologic functions, such as motility, fluid secretion, and blood flow, by secreting hormones and neurotransmitters. More than 20 different EE cell types are recognized, and they differ in their gut location and type of hormones secreted (8). The EE cells differ from most other epithelial cells, such as parietal cell or enterocytes, that are programmed to produce acid and digestive enzymes to digest food components into their simplest, constituent units, namely, sugars, fatty acids, and amino acids.
Some examples of specific nutrient sensing in EE cells include their responses to carbohydrate, lipids, and protein. Glucose absorption from the intestine occurs through the sodium/glucose cotransporter 1 (SGLT1) expressed mainly on absorptive enterocytes. In vivo animal studies showed that the SGLT1 transporter is up-regulated by the presence of glucose as well as by nonmetabolizable glucose analogs in the intestinal lumen (8, 9). The mechanism by which glucose increases the expression of SGLT1 in enterocytes is thought to involve EE cells because they also express the same taste receptors (T1R2 + T1R3) present in taste buds (see Fig. 47.1). Glucose also activates the EE cell release of incretin hormones involved in insulin secretion and peripheral glucose uptake, namely, glucosedependent insulinotropic peptide (GIP) and the glucagonlike peptides, GLP-1 and GLP-2. The current theory is that glucose-dependent activation of T1R in EE cells triggers the release of hormones that eventually lead to increased SGLT1 expression and glucose uptake. Luminal glucose also can activate EE cell release of 5-hydroxytryptamine (5-HT) or serotonin, which regulates gastric emptying and pancreatic exocrine and intestinal fluid secretion by interaction with vagal afferent neural circuits (8, 10).
The EE cells also respond to luminal lipids by secretion of cholecystokinin (CCK), which regulates several physiologic GI functions and appetite by activation of vagal nerves (8). The mechanism for EE cell sensing has been ascribed to the presence of several seven-transmembrane GPCRs that recognize fatty acids, namely, GPR120, FFAR1, FFAR2, and FFAR3. These cell receptors are expressed on EE cells that colocalize with GLP-1 and peptide YY (PYY) (11).
Products of protein hydrolysis also activate release of CCK by EE cells, with physiologic effects similar to those of lipids (10). Peptides are taken up by the intestinal enterocyte apical membrane by peptide transporter 1 (PepT1), a cotransporter specific for dipeptides and tripeptides. Studies suggested that peptidomimetic compounds specific for PepT1 induce CCK release and the expected inhibition of gastric motility. Emerging reports indicate that additional amino acid sensing receptors, including T1R3, metabotropic glutamate receptor 1 to 4, and calcium-sensing receptor (CaSr), are also expressed on EE cells and other epithelial cells in the gut (12, 13). The physiologic significance of these cell receptors in nutrient sensing awaits further study.
INTRACELLULAR NUTRIENT SENSORS
Glucose, Amino Acid, and Fatty Acid Sensing
Once nutrients are absorbed from the intestine into the bloodstream, they are sensed by somatic cells through various cellular mechanisms. Several intracellular or nuclear receptors are regulated by changes in the cellular availability of nutrients such as glucose, amino acids, and fatty acids.
Mechanisms Related to Carbohydrate-Responsive Element-Binding Protein and Liver X Receptor
An important cellular glucose sensing mechanism involves the transcription factor carbohydrate-response elementbinding protein (ChREBP), which is activated in response to increased cellular glucose concentrations (see Fig. 47.1) (14, 15). ChREBP is mainly expressed not only in the liver but also in other glucose-responsive tissues such as adipose, brain, and pancreas. In fasting conditions of low glucose concentrations, ChREBP is located in the cytosol in its phosphorylated form associated with the 14-3-3 protein. After a meal, however, the increased cellular influx of glucose results in increased production of xylulose 5-phosphate (X5P) through the pentose phosphate pathway. Increased cellular X5P concentration leads to activation of protein phosphatase 2A, which dephosphorylates ChREBP and enables its translocation in the nucleus.
Once in the nucleus, ChREBP interacts with a binding partner, Max-like protein X, which then binds to the carbohydrate-response element of multiple target genes and this increases their transcription. Many of the target genes activated by ChREBP are enzymes involved in lipogenesis and glucose metabolism. Another transcription factor reported to bind glucose is the nuclear receptor liver X receptor (LXR), whose primary ligands are oxysterols such as cholesterol (16). Ligand activation of LXR by oxysterols induces heterodimerization with retinoid X receptor and binding of target gene promoter sequences, processes that lead to activation of lipogenesis in liver and adipose tissue. The relative importance of ChREBP compared with LXR in glucose sensing has not been identified (17).
Once in the nucleus, ChREBP interacts with a binding partner, Max-like protein X, which then binds to the carbohydrate-response element of multiple target genes and this increases their transcription. Many of the target genes activated by ChREBP are enzymes involved in lipogenesis and glucose metabolism. Another transcription factor reported to bind glucose is the nuclear receptor liver X receptor (LXR), whose primary ligands are oxysterols such as cholesterol (16). Ligand activation of LXR by oxysterols induces heterodimerization with retinoid X receptor and binding of target gene promoter sequences, processes that lead to activation of lipogenesis in liver and adipose tissue. The relative importance of ChREBP compared with LXR in glucose sensing has not been identified (17).
Mechanisms Related to Activating Transcription Factor 4
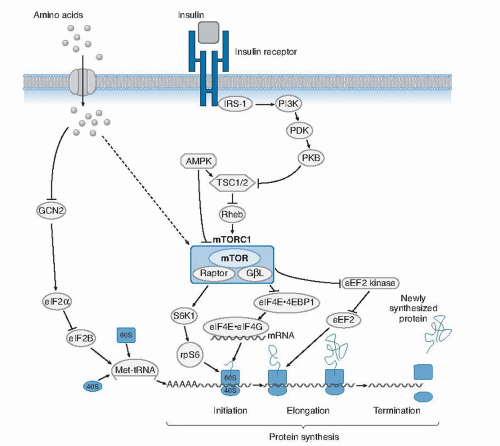
A key cellular mechanism for sensing amino acids, especially in conditions of amino acid deficiency or imbalance, involves stimulation of activating transcription factor 4 (ATF4) (see Fig. 47.1) (18). In conditions of food deprivation or dietary protein restriction, the level of activated or charged transfer RNA (tRNA) bound to amino acids decreases. This results in an increase in uncharged tRNAs, which bind to the general control non-derepressible 2 (GCN2) kinase and, in turn, increase the phosphorylation of the eukaryotic initiation factor 2α (eIF2α) (Fig. 47.2). The phosphorylated form of eIF2α suppresses general protein synthesis by inhibiting translation initiation factor, eIF2B, which is critical for ribosomal assembly.
Paradoxically, the amino acid-limiting conditions alter ribosomal processing and result in increased translation of ATF4, which then binds to a conserved promoter region in many genes involved in amino acid transport (cationic amino acid transporter [CAT-1] and system A sodiumdependent neutral amino acid transporter [SNAT2]), metabolism (asparagine synthetase [ASNS]), and cell death (C/EBP homologous protein [CHOP] and tribbles homolog 3 [TRB3]). The ATF4 signaling pathway senses amino acid stress and functions as a counterpart to the mammalian target of rapamycin (mTOR) pathway (discussed later) that responds to amino acid sufficiency to promote cell anabolism and growth.
Paradoxically, the amino acid-limiting conditions alter ribosomal processing and result in increased translation of ATF4, which then binds to a conserved promoter region in many genes involved in amino acid transport (cationic amino acid transporter [CAT-1] and system A sodiumdependent neutral amino acid transporter [SNAT2]), metabolism (asparagine synthetase [ASNS]), and cell death (C/EBP homologous protein [CHOP] and tribbles homolog 3 [TRB3]). The ATF4 signaling pathway senses amino acid stress and functions as a counterpart to the mammalian target of rapamycin (mTOR) pathway (discussed later) that responds to amino acid sufficiency to promote cell anabolism and growth.
Stay updated, free articles. Join our Telegram channel

Full access? Get Clinical Tree