18.1 INTRODUCTION TO HIV
There are two types of human immunodeficiency virus (HIV-1 and HIV-2), which each evolved from a different simian immunodeficiency virus (SIV). Both HIVs emerged in the late twentieth century. HIV infection damages the immune system, leaving the body susceptible to infection with a wide range of bacteria, viruses, fungi, and protozoa. This condition is called acquired immune deficiency syndrome (AIDS). It should be noted that the word “virus” in the phrase “HIV virus” is superfluous and that scientists use the abbreviation “AIDS”, not “Aids”!
HIV continues to spread, with millions of new infections each year. AIDS continues to kill large numbers of people, and has become the fourth biggest cause of mortality in the world. The magnitude of this problem has resulted in the allocation of huge resources to the study of these viruses, major objectives being the development of anti-viral drugs and a vaccine. So far there has been some success in achieving the first of these objectives.
HIV-1 is largely responsible for the AIDS pandemic, while HIV-2 is mainly restricted to West Africa. Consequently the emphasis of this chapter is on HIV-1, as it has been studied more intensively than HIV-2.
18.2 HIV VIRION
The virion has the general characteristics of retroviruses described in the previous chapter but, in contrast to most retroviruses, the capsid is cone-shaped (Figure 18.1), with a diameter of 40–60 nm at the wide end and about 20 nm at the narrow end. Most of the capsid protein is present as hexamers, with five pentamers at the narrow end of the cone and seven pentamers at the wide end of the cone. Generally, there is one capsid per virion, though virions with two or more capsids have been reported.
Figure 18.1 HIV virion. (a) Virion components. IN: integrase. NC: nucleocapsid protein. RT: reverse transcriptase. The TM and SU glycoproteins indicated are those of HIV-1 (gp41 and gp120). (c) Capsid model, showing protein hexamers in green and pentamers in red.
Sources: (b) Grünewald and Cyrklaff (2006) Current Opinion in Microbiology, 9, 437. (c) Ganser-Pornillos, Yeager, and Sundquist (2008) Current Opinion in Structural Biology, 18, 203. (b) and (c) reproduced by permission of Elsevier Limited and the authors
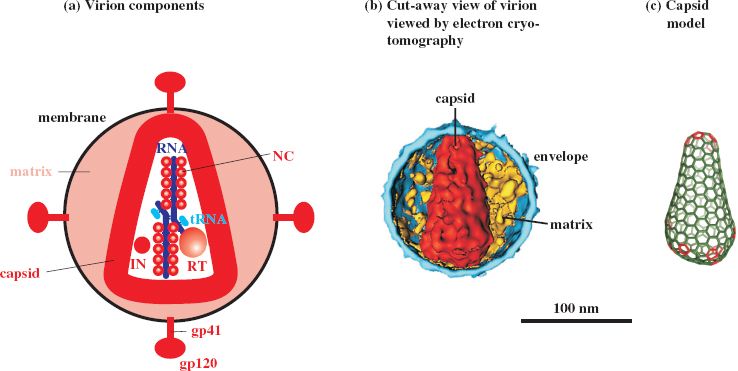
The diameter of the HIV virion measured in negatively stained preparations is in the range 80–110 nm, while results from cryo-electron microscopy are at the upper end of this range or greater.
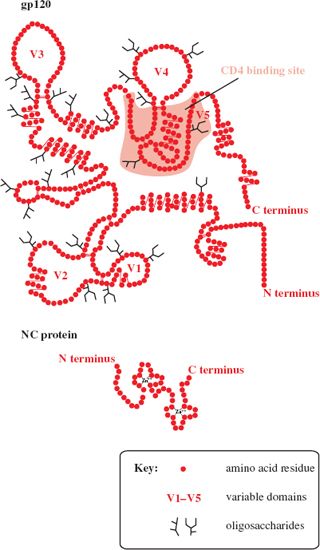
The TM and SU proteins of HIV-1 have approximate molecular weights of 41 kD and 120 kD, respectively, and are named gp41 and gp120 (gp = glycoprotein). gp120 is heavily glycosylated and has five domains near the surface that are highly variable (V1–V5; Figure 18.2). The C terminus of gp41 is inside the virion, where it is bound to the MA protein. Spikes can be seen at the surface of virions in electron micrographs, with an average of 14 spikes on each virion. Each spike is a gp41–gp120 trimer. The equivalent glycoproteins in HIV-2 (gp38 and gp130) are unrelated to those of HIV-1, whereas most of the internal proteins of the two viruses are related. The NC proteins are similar to those of many other retroviruses; they have zinc fingers (Figure 18.2) and are highly basic (29% of the amino acid residues of HIV-1 NC protein are basic).
Figure 18.2 Structures of HIV-1 gp120 and NC protein. gp120 has five domains that are highly variable (V1–V5). The NC protein has two zinc fingers.
As well as the standard retrovirus proteins, the HIV-1 virion also contains the following virus proteins: Nef, Vpr, Vif, p1, p2, p6, and p6∗. The presence of host proteins has also been reported, including major histocompatibility complex class II proteins associated with the envelope, and cyclophilin A associated with the capsid.
18.3 HIV GENOME
HIV-1 and HIV-2 have genomes about 9.3 kb in length. In addition to the standard retroviral genes gag, pol, and env (Figure 17.1) there are auxiliary genes, so the viruses are classed as complex retroviruses. The proteins encoded by the auxiliary genes can interact with a wide range of HIV and cell components. Each protein has multiple roles, and together they control virus gene expression, modify the host’s immune response, and are involved in transporting virus components within the cell.
The organization of the HIV-1 genome is shown in Figure 18.3. All three reading frames are used and there is extensive overlapping; for example, part of vpu in frame 2 overlaps env in frame 3. The sequences for tat and rev are split, the functional sequences being formed when the transcripts are spliced.
HIV-2 has similar genes to HIV-1, except that it has no vpu gene, but it has a vpx gene, which is related to vpr.
18.4 HIV-1 REPLICATION
A general description of the retrovirus replication cycle was given in the previous chapter. Here we shall concentrate on details specific to HIV-1.
18.4.1 Attachment and entry
The cell receptor for HIV-1 is CD4 (Figure 18.4), which is a member of the immunoglobulin superfamily of molecules. CD4 is found on several cell types, including helper T cells and some macrophages; CD4 T cells are the main host cells. Attachment of the virion occurs when a site on gp120 (Figure 18.2) recognizes a site on the outer domain of CD4.
Figure 18.4 HIV-1 attachment and entry. The receptor is CD4; each of the loops represents an immunoglobulin-like domain, three of which are stabilized by disulfide bonds. The co-receptor is a chemokine receptor. Fusion of the virion and cell membranes releases a structure that forms a reverse transcription complex, consisting of the virus genome, tRNA, and several proteins, including Vpr.
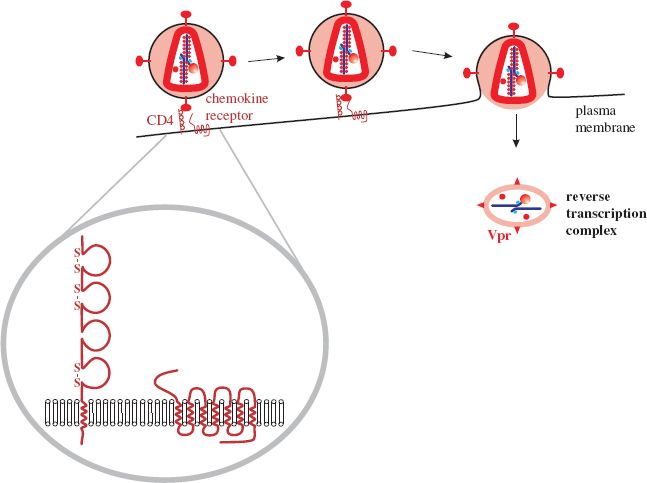
As well as binding to receptor molecules gp120 must also attach to a co-receptor on the cell surface. The molecules that act as co-receptors have seven transmembrane domains and are chemokine receptors. During immune responses they bind chemokines and these interactions control leukocyte trafficking and T cell differentiation. Most chemokines fall into one of two major classes, determined by the arrangement of cysteine residues near the N terminus: C—C and C—X—C, where C = cysteine and X = any amino acid. The chemokine receptors are designated CCR and CXCR, respectively. A number of these molecules on T cells act as co-receptors for HIV-1, particularly CCR5 and CXCR4 (Table 18.1).
Table 18.1 Co-receptors for HIV-1 strains and categories of T cell infected.
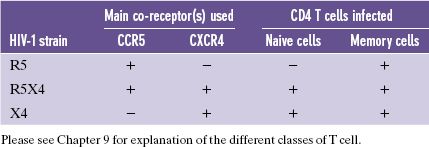
Most HIV-1 strains use CCR5 and are known as R5 strains. It is interesting to note that in some individuals who have had multiple exposures to the virus, but have not become infected, there is a 32-nucleotide deletion in the CCR5 gene. Individuals who are heterozygous for this mutation are less susceptible to infection with HIV-1, while those who are homozygous express no CCR5 on their cells and are highly resistant. The mutation is found mainly in Europeans.
HIV-1 strains that use CXCR4 as a co-receptor are known as X4 strains, and there are some HIV-1 strains (R5X4 strains) that can use either co-receptor. R5 strains do not infect naive T cells, but all three strains infect memory T cells.
The interaction of gp120 with the receptor and co-receptor results in a dramatic rearrangement of gp41, which proceeds to fuse the membranes of the virion and the cell. The matrix and nucleocapsid are released into the cytoplasm and develop into the reverse transcription complex, which contains the MA, Vpr, RT, and IN proteins, as well as the virus genome and tRNA (Figure 18.4).
18.4.2 Reverse transcription and transport to the nucleus
The reverse transcription complex associates rapidly with microtubules (Figure 18.5). Reverse transcription is primed by tRNAlys3, and proceeds as outlined in Section 17.3.2.
Figure 18.5 HIV-1 reverse transcription and integration of the provirus. A DNA flap is formed during reverse transcription. There is evidence that the pre-integration complex is transported toward the nucleus via the microtubule network.
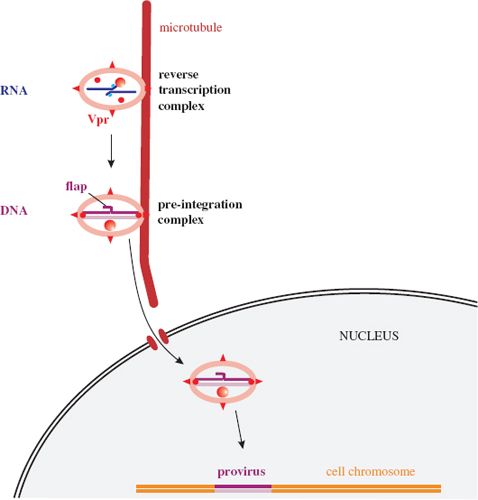
Whereas the proviruses of most retroviruses are entirely dsDNA, those of HIV and other lentiviruses have a short triple-stranded sequence known as a central DNA flap (Figure 18.5). This comes about because there are two initiation sites for (+) DNA synthesis; as well as the polypurine tract (PPT) toward the 3′ end of the virus genome, there is also a central PPT within the pol region. Synthesis of the (+) DNA initiated at the 3′ PPT stops soon after reaching the (+) DNA initiated at the central PPT, resulting in a short overlapping DNA sequence. This DNA flap plays a vital role in the early stages of infection.
After reverse transcription has been completed, the pre-integration complex, which contains host proteins as well as virus proteins, is transported into the nucleus. There is evidence that transport toward the nucleus takes place on the microtubule network. As discussed in Chapter 17, most retroviruses can productively infect only if there is breakdown of the nuclear membranes. The pre-integration complex of HIV, however, can enter an intact nucleus, such as that of a resting T cell or a macrophage, and is presumably transported through a nuclear pore.
The viral integrase and cell enzymes are involved in the integration of the provirus into a cell chromosome; cell enzymes remove the DNA flap and repair the gap. There is evidence that integration of the provirus in a resting memory CD4 T cell may result in a latent infection. Latently infected cells can provide a reservoir of infection that is significant for the survival of the virus in individuals receiving anti-retroviral drug therapy. In many cells, though, provirus integration is the prelude to a productive infection in which two phases of gene expression can be distinguished.
18.4.3 Early gene expression
Transcription is initiated after cell transcription factors bind to promoter and enhancer sequences in the U3 region of the upstream LTR. This region has binding sites for a number of cell transcription factors, including AP-1, NF-κB, and Sp-1 (Figure 6.4). Transcription is terminated in the downstream LTR; the polyadenylation signal AATAAA is in the R region and transcripts are polyadenylated at the R–U5 junction.
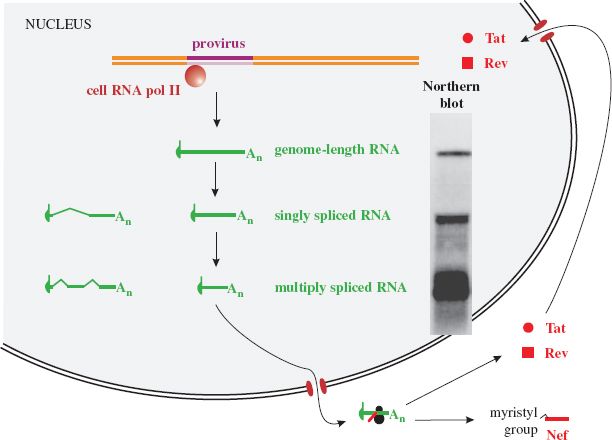
Many of the transcripts are spliced, resulting in three size classes of virus transcript. These can be detected in infected cells using northern blotting (Figure 18.6). The largest RNAs are genome length (about 9.3 kb), while the other two size classes are each made up of a number of mRNA species that have undergone splicing; mRNAs that have been spliced once are around 4.5 kb, while mRNAs that have undergone two or more splicing events (“multiply spliced” transcripts) are around 2 kb. The virus genome has a number of splice donor sites (one is indicated in Figure 18.3) and acceptor sites; these enable splicing events that result in more than 40 mRNA species.
Figure 18.6 HIV-1 early gene expression. Genome-length RNA is transcribed then much of it is spliced, giving rise to two further size classes of RNA that can be detected in northern blots of RNA from infected cells. Early in infection most of the RNA is multiply spliced and is transported to the cytoplasm, where the Tat, Rev, and Nef proteins are translated. Nef is myristylated and performs a number of roles in the cytoplasm, while Tat and Rev are transported to the nucleus.
Source: Northern blot from Malim et al. (1990) Cell, 60, 675; reproduced by permission of Elsevier Limited and the authors.
Stay updated, free articles. Join our Telegram channel

Full access? Get Clinical Tree