How Cells Function: Fundamental Concepts of Molecular Biology*
Molecular biology is a branch of the biologic sciences that attempts to explain life and its manifestations as a series of chemical and physical reactions. The critical event that led to the development of this new science was the discovery of the fundamental structure of deoxyribonucleic acid (DNA) by Watson and Crick in 1953. Few prior developments in biology have contributed so much and so rapidly to our understanding of the many fundamental aspects of cell function and genetics. Although, so far, the impact of molecular biology on diagnostic cytology has been relatively modest, this may change in the future. Therefore, some of the fundamental principles of this new science are briefly summarized. The main purpose of this review is to describe the events in DNA replication, transcription, and translation of genetic messages; to clarify the new terminology that has entered into the scientific vocabulary since 1953; and to explain the techniques that are currently used to probe the functions of the cell. It is hoped that this review will enable the reader to follow future developments in this stillexpanding field of knowledge. Of necessity, this summary touches upon only selected aspects of molecular biology, representing a personal choice of topics that, in the judgment of the writer, are likely to contribute to diagnostic cytology. For reasons of economy of space, with a very few exceptions, the names of the many investigators who contributed
to this knowledge are not used in this text. Readers are referred to other sources listed in the bibliography for a more detailed record of individual contributors and additional information on specific technical aspects of this challenging field.
to this knowledge are not used in this text. Readers are referred to other sources listed in the bibliography for a more detailed record of individual contributors and additional information on specific technical aspects of this challenging field.
Molecular biology is easily understood because it is logical and based on the simple principles of organic chemistry. Hence, basic knowledge of organic chemistry is necessary to understand the narrative. Every attempt has been made to tell the story in a simple language.
THE CELL AS A FACTORY
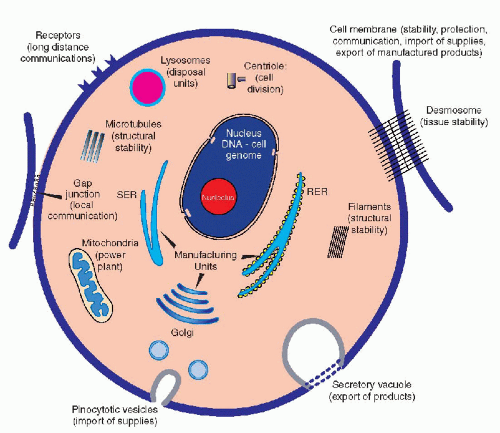
Although the main morphologic components of the cell have been identified by light and electron microscopy (see Chap. 2), until 5 decades ago, the understanding of the mechanisms governing cell function has remained elusive and a matter for conjecture. Molecular biology has now shed light on some of these mechanisms, although, at the time of this writing (2004), much remains to be discovered. The living cell is best conceived as a self-contained miniature factory that must fulfill a number of essential requirements necessary to manufacture products, either for its own use or for export (Fig. 3-1). A cell is a three-dimensional structure contained within the cell membrane, which is a highly sophisticated, flexible structure (see Chap. 2). The membrane not only protects the cell from possible hostile elements or environmentally unfavorable conditions, but it is also capable of selective intake of materials that are important and necessary to the survival of the cell; this latter property is vested in specialized molecular sites: the membrane receptors (see Chap. 2). The cell exports finished products by using intricate mechanisms in which the cell membrane is an active participant. The membrane is also provided with a series of devices, such as cell junctions, which allow the cell to live in harmony and to communicate with its neighbors.
The cell is constructed in a sturdy fashion, thanks to the cell skeleton composed of microfilaments, intermediate filaments, and microtubules (see Chap. 2). The cell is capable of producing the components of its own skeleton and of regulating their functions. The energy needs of the cell are provided by the metabolism of foodstuffs, mainly sugars and fats, interacting with the energy-producing systems, adenosine 5%-triphosphate (ATP), vested primarily in the mitochondria. The machinery that allows the cell to manufacture or synthesize products for its own use or for export, mainly a broad variety of proteins, is vested in the system of cytoplasmic membranes, the smooth and rough endoplasmic reticulum, and in the ribosomes (see Chap. 2). Disposal of useless or toxic products is vested in the system of lysosomes and related organelles. As a signal advantage of most cells over a manmade factory, the cell is
provided with a system of reproduction in its own image, in the form of cell division or mitosis (see Chap. 4). Thus, aged and inadequately functioning cells may be replaced by daughter cells, which ensure the continuity of the cell lineage, hence of the tissue, and ultimately of the species. The equilibrium among cells is also maintained by a mechanism of elimination of unwanted or unnecessary cells by a process known as apoptosis, or programmed cell death. Apoptosis plays an important role during embryonal development, wherein unnecessary cells are eliminated in favor of cells that are needed for construction of tissues or organs with a definite function. Apoptosis also occurs in adult organisms and may play an important role in cancer. The mechanisms of apoptosis are complex and consist of a cascade of events, involving the mitochondria and the nuclear DNA, discussed at length in Chapter 6.
provided with a system of reproduction in its own image, in the form of cell division or mitosis (see Chap. 4). Thus, aged and inadequately functioning cells may be replaced by daughter cells, which ensure the continuity of the cell lineage, hence of the tissue, and ultimately of the species. The equilibrium among cells is also maintained by a mechanism of elimination of unwanted or unnecessary cells by a process known as apoptosis, or programmed cell death. Apoptosis plays an important role during embryonal development, wherein unnecessary cells are eliminated in favor of cells that are needed for construction of tissues or organs with a definite function. Apoptosis also occurs in adult organisms and may play an important role in cancer. The mechanisms of apoptosis are complex and consist of a cascade of events, involving the mitochondria and the nuclear DNA, discussed at length in Chapter 6.
It is quite evident from this brief summary that a highly sophisticated system of organization, which will coordinate its many different functions, must exist within each cell. Furthermore, within multicellular organisms, these functions vary remarkably from cell to cell and from tissue to tissue; hence, they must be governed by a flexible mechanism of control. The dominant role in the organization of the cell function is vested in the DNA, located in the cell’s nucleus. The mechanisms of biochemical activities directed by DNA and the interaction of molecules encoded therein is the subject of this summary.
DEOXYRIBONUCLEIC ACID (DNA)
Background
The recognition of the microscopic and ultrastructural features of cells and their fundamental components, such as the nucleus, the cytoplasm with its organelles, and the cell membrane, all described in Chapter 2, shed little light on the manner in which cells function. The key questions were: How does a cell reproduce itself in its own image? How are the genetic characteristics of cells inherited, transmitted, and modified? How does a cell function as a harmonious entity within the framework of a multicellular organism?
The facts available to the investigators during the 100 years after the initial observations on cell structure were few and difficult to reconcile. The developments in organic chemistry during the 19th century documented that the cells are made up of the same elements as other organic matter, namely, carbon, hydrogen, oxygen, nitrogen, phosphorous, calcium, sulfur, and very small amounts of some other inorganic elements. Perhaps the most critical discovery was the synthesis of urea by Wöhler in 1828. Soon, a number of other organic compounds, such as various proteins, fats and sugars, were identified in cells. Of special significance for molecular biology was the observation that all proteins are composed of the same 20 essential amino acids. A further important observation was that most enzymes, hence substances responsible for the execution of many chemical reactions, were also proteins. The cell ceased to be a chemical mystery, but it remained a functional puzzle.
The observations by the Czech monk, Gregor Mendel (or Mendl), who first set down the laws governing dominant and recessive genetic inheritance by simple observations on garden peas, opened yet another pathway to molecular biology. Was there any possible link between biochemistry and genetics? The phenomenon of mitosis, or cell division, and the presence of chromosomes were first observed about 1850, apparently by one of the founding fathers of contemporary pathology, Rudolf Virchow. Several other 19th-century observers described chromosomes in some detail and speculated on their possible role in genetic inheritance, but, again, there was no obvious way to reconcile the chromosomes with the genetic and biochemical data.
In 1869, a Swiss biochemist, Miescher, isolated a substance from the nuclei of cells from the thymus of calves, named thymonucleic acid, and since renamed deoxyribonucleic acid or DNA. The relationship between DNA and the principles of genetic inheritance, as defined by Mendel, was not apparent for almost a century. A hint linking the chromosomes with the “thymonucleic” acid was provided by Feulgen and Rossenbach, who, in 1924, devised a DNA-specific staining reaction, which is known today as the Feulgen stain. It could be shown that chromosomes stained intensely with this stain (see Fig. 2-28). Interestingly, in the 1930s, the Swedish pioneer of cytochemistry, Torbjörn Caspersson, suggested that thymonucleic acid could be the substance responsible for genetic events in the cell.
It was not, however, until 1944 that Avery, MacCarty, and MacLeod, working at the Rockefeller Institute in New York City, described a series of experiments documenting that DNA was the molecule responsible for morphologic changes in the bacterium, Diplococcus pneumoniae, thus providing firm underpinning to the principle that the genetic function was vested in this compound. The universal truth of this discovery was not apparent for several more years, particularly because bacterial DNA does not form chromosomes. The understanding of the mechanisms of the function of DNA had to await the discovery of the fundamental structure of this molecule by Watson and Crick in 1953. For a recent review of these events, see Pennisi (2003).
Structure
DNA was once described as a “fat, cigar-smoking molecule that orders other molecules around.” In fact, the molecule of DNA is central to all events occurring within the cell. In bacteria and other relatively simple organisms not provided with a nucleus (prokaryotes), the DNA is present in the cytoplasm. In higher organisms (eukaryotes), most of the DNA is located within the nucleus of the cell. In a nondividing cell, the DNA was thought to be diffusely distributed within the nucleus. Recent investigations, however, strongly suggest that even in the nondividing cells, the chromosomes retain their identity and occupy specific territories within the nucleus (Koss, 1998). For further details of the nuclear structure, see Chapter 2. During cell division, the DNA is condensed into visible chromosomes (see Chap. 4). Small amounts of DNA are also present in other cell organelles, mainly in the mitochondria; hence, the suggestion
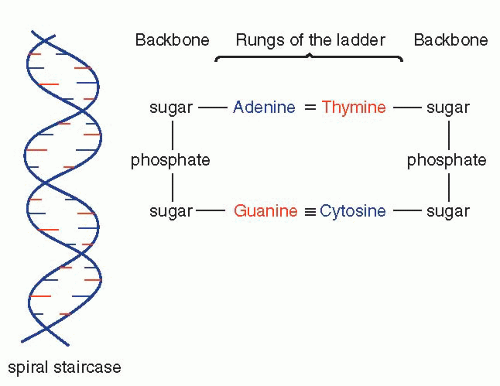
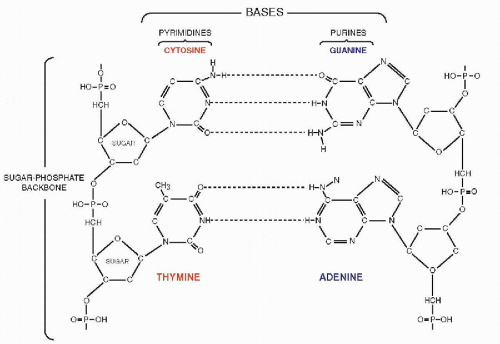
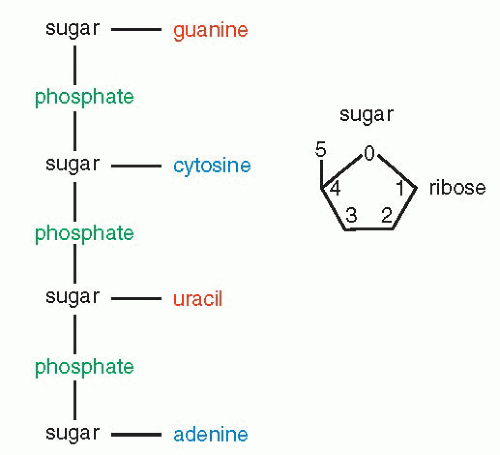
that mitochondria represent previously independent bacterial organisms that found it advantageous to live in symbiosis with cells (see Chap. 2). To understand how DNA performs the many essential functions, it is important to describe its structure. DNA forms the well-known double helix, which can be best compared to an ascending spiral staircase or a twisted ladder (Fig. 3-2). The staircase has a supporting external structure, or backbone, composed of molecules of a pentose sugar, deoxyribose, bound to one another by a molecule of phosphate. This external support structure of the staircase is organized in a highly specific fashion: the organic rings of the sugar molecules are alternately attached to the phosphate by their 5′ and 3′ carbon molecules* (Fig. 3-3). This construction is fundamental to the understanding of the synthesis of nucleic acids, which always proceeds from the 5′ to the 3′ end, by addition of sugar molecules in the 3′ position. The steps of the staircase (or rungs of the ladder) are formed by matching molecules of purine and pyrimidine bases, each attached to a molecule of the sugar, deoxyribose, in the backbone of the molecule (Fig. 3-4; see Fig. 3-2). The purines are adenine (A) and guanine (G); the pyrimidines are thymine (T) and cytosine (C). It has been known since the 1940s, thanks to the contributions of the chemist, Chargaff, that in all DNA molecules, regardless of species of origin, the proportions of adenine and thymine on the one hand, and of guanine and cytosine on the other hand, were constant. This information, combined with data from x-ray crystallography of purified molecules of DNA, allowed Watson and Crick to construct their model of the DNA molecule. In it, the purine, adenine, and the pyrimidine, thymine (the A-T bond), and cytosine and guanine (the C-G bond) are always bound to each other. The triple C-G bond is stronger than the double A-T bond (see Figs. 3-2 and 3-4). This relationship of purines and pyrimidines is immutable, except for the replacement of thymine by uracil (U) in RNA (see below), and is the basis of all subsequent technical developments in the identification of matching fragments of nucleic acids (see below). The term base pairs (bp) is frequently used to define one matching pair of nucleotides and to define the length of a segment of double-stranded DNA. Thus, a DNA molecule may be composed of many thousands of base pairs. It is of critical importance to realize that the sequence of the purine-pyrimidine base pairs varies significantly, in keeping with the encoding of the genetic message, as will be set forth below.
that mitochondria represent previously independent bacterial organisms that found it advantageous to live in symbiosis with cells (see Chap. 2). To understand how DNA performs the many essential functions, it is important to describe its structure. DNA forms the well-known double helix, which can be best compared to an ascending spiral staircase or a twisted ladder (Fig. 3-2). The staircase has a supporting external structure, or backbone, composed of molecules of a pentose sugar, deoxyribose, bound to one another by a molecule of phosphate. This external support structure of the staircase is organized in a highly specific fashion: the organic rings of the sugar molecules are alternately attached to the phosphate by their 5′ and 3′ carbon molecules* (Fig. 3-3). This construction is fundamental to the understanding of the synthesis of nucleic acids, which always proceeds from the 5′ to the 3′ end, by addition of sugar molecules in the 3′ position. The steps of the staircase (or rungs of the ladder) are formed by matching molecules of purine and pyrimidine bases, each attached to a molecule of the sugar, deoxyribose, in the backbone of the molecule (Fig. 3-4; see Fig. 3-2). The purines are adenine (A) and guanine (G); the pyrimidines are thymine (T) and cytosine (C). It has been known since the 1940s, thanks to the contributions of the chemist, Chargaff, that in all DNA molecules, regardless of species of origin, the proportions of adenine and thymine on the one hand, and of guanine and cytosine on the other hand, were constant. This information, combined with data from x-ray crystallography of purified molecules of DNA, allowed Watson and Crick to construct their model of the DNA molecule. In it, the purine, adenine, and the pyrimidine, thymine (the A-T bond), and cytosine and guanine (the C-G bond) are always bound to each other. The triple C-G bond is stronger than the double A-T bond (see Figs. 3-2 and 3-4). This relationship of purines and pyrimidines is immutable, except for the replacement of thymine by uracil (U) in RNA (see below), and is the basis of all subsequent technical developments in the identification of matching fragments of nucleic acids (see below). The term base pairs (bp) is frequently used to define one matching pair of nucleotides and to define the length of a segment of double-stranded DNA. Thus, a DNA molecule may be composed of many thousands of base pairs. It is of critical importance to realize that the sequence of the purine-pyrimidine base pairs varies significantly, in keeping with the encoding of the genetic message, as will be set forth below.
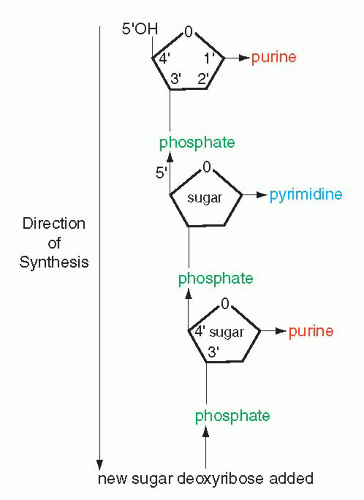 Figure 3-3 Schematic representation of the backbone of DNA and the direction of synthesis from 5′ to 3′, indicating positions of carbons in the molecules of sugar. |
Packaging
DNA is an enormous molecule. If fully unwrapped, it measures about 2 meters in length (but only 2 nm in diameter) in each single human nucleus. Each of the 46 individual human chromosomes contains from 40 to 500 million base pairs and their DNA is, therefore, of variable length, but still averages about 3 cm. It is evident, therefore, that to fit this gigantic molecule into a nucleus measuring from 7 to 10 μm in diameter, it must be folded many times. The DNA is wrapped around nucleosomes, which are cylindrical structures, composed of proteins known as histones (see Fig. 4-5). This reduces the length of the molecule significantly. Further reduction of the molecule is still required, and it is assumed that DNA forms multiple coils and folds to form a compact structure that fits into the space reserved for the nucleus. An apt comparison is with a wet towel that is twisted to rid it of water and then folded and refolded to form a compact ball. The interested reader is referred to a delightful book by Calladine and Drew (1997) that explains in a simple fashion what is known today about packaging of DNA. Be it as it may, individual chromosomes are
composed of multiple coils of DNA, as shown in Figure 4-5 and discussed at some length in Chapter 4.
composed of multiple coils of DNA, as shown in Figure 4-5 and discussed at some length in Chapter 4.
Replication
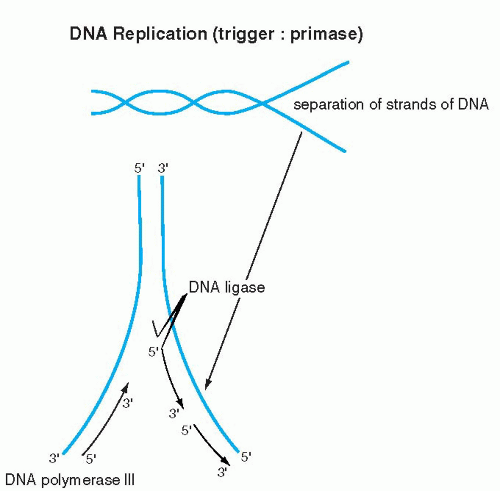
The elegance and simplicity of the structure of the double helix resolved the secret of inheritance of genetic material. Because the double helix is constructed of two reciprocal, matching molecules, it was evident to Watson and Crick that DNA replication can proceed in its own image: the double helix can be compared to a zipper, composed of two corresponding half-zippers. Each half of the zipper, or one strand of DNA, serves as a template for the formation of a mirror image, complementary strand of DNA (Fig. 3-5). Hence, the first event in DNA replication must be the separation of the two strands forming the double helix. The precise mechanism of strand separation is still not fully understood, although the enzyme primase plays an important role. A further complication in the full understanding of the mechanisms of DNA replication is that the DNA molecule is wrapped around nucleosomes (see above). How the nucleosomal DNA is unwrapped and replicated, or for that matter transcribed (see below), is not fully understood as yet.
The synthesis of the new strand, governed by enzymes known as DNA polymerases, follows the fundamental principle of A-T and G-C pairing bonds and the principle of the 5′-to-3′ direction of synthesis, as described above. Because the two DNA strands are reciprocal, the synthesis on one strand is continuous and proceeds without interruption in the 5′-to-3′ direction. The synthesis on the other strand also follows the 5′-to-3′ rule but must proceed in the opposite direction; hence, it is discontinuous (Fig. 3-6). The segments of DNA created in the discontinuous manner are spliced together by an enzyme, ligase. When both strands of DNA (half-zippers) are duplicated, two identical molecules (fullzippers) of DNA are created. This fundamental basis of DNA replication permits the daughter cells to inherit all the characteristics of the mother cell that are vested in the DNA. Replication of DNA takes place during a well-defined period in a cell’s life, the synthesis phase or S-phase of the cell cycle, before the onset of cell division (mitosis) (see Chap. 4). By the time the cell enters the mitotic division, the DNA, in the form of chromosomes; is already duplicated. Each chromosome is composed of two identical mirror-image DNA segments (chromatids), bound together by a centromere (see Fig. 4-2). It is evident that the mechanism of DNA replication is activated before mitosis, when the chromosomal DNA is not visible under the light microscope, because the chromosomes are markedly elongated.
The exact sequence of events leading to the entry of the cell into the mitotic cycle is still under investigation and may be influenced by extracellular signals (see review by Cook, 1999). Whatever the mechanism, a family of proteins, cyclins, causes the resting cell to enter and progress through the phases of the cell division. For a review of cyclins, see the article by Darzynkiewicz et al (1996) and Chapter 4.
It is also known that the replication of the chromosomes
is not synchronous and that some of them replicate early and others replicate late. It has been proposed that those genes common to all cells that ensure the fundamental cell functions and “housekeeping” chores, replicate during the first, early part of the S-phase, whereas the tissue-specific genes replicate late. During other phases of the cell cycle, the mechanism of DNA replication is either inactive or markedly reduced.
is not synchronous and that some of them replicate early and others replicate late. It has been proposed that those genes common to all cells that ensure the fundamental cell functions and “housekeeping” chores, replicate during the first, early part of the S-phase, whereas the tissue-specific genes replicate late. During other phases of the cell cycle, the mechanism of DNA replication is either inactive or markedly reduced.
 Figure 3-5 The DNA molecule and its manner of replication. Each base pair and its respective sugar-phosphate helix comes apart and induces synthesis of its complementary chain. |
If one considers that during the lifetime of the human organism, DNA replication occurs billions of times and that even single errors of replication affecting critical segments of DNA may result in serious genetic damage that may lead to clinical disorders (see below), it is evident that efficient mechanisms of replication control must exist that will eliminate or neutralize such mistakes. Work on bacteria suggests that there are at least three controlling steps in DNA replication: selection of the appropriate nucleotide by DNA polymerases; recognition of the faulty structure by another enzyme; and finally, the repair of the damage. In eukaryotic cells, the molecule p53, which has been named “the guardian of the genome,” appears to play a critical role in preventing replication errors prior to mitosis. As discussed in Chapter 6, cells that fail to achieve DNA repair will be eliminated by the complex mechanism of apoptosis. Regardless of the technical details, it is quite evident that these control mechanisms of DNA replication in multicellular organisms are very effective.
Transcription
Once the fundamental structure of DNA became known, attention turned to the manner in which this molecule governed the events in the cell. There were two fundamental questions to be answered: How were the messages inscribed in the DNA molecule (i.e., how was the genetic code constructed?) and how were they executed? It became quite evident that the gigantic molecule of DNA could not be directly involved in cell function, particularly in the formation of the enzymes and other essential molecules. Furthermore, it had been known that protein synthesis takes place in the cytoplasm and not in the nucleus; hence, it became clear that an intermediate molecule or molecules had to exist to transmit the messages from the nucleus to the cytoplasm (see Fig. 3-8A). The best candidate for this function was RNA. RNAs, or the ribose nucleic acids, were analyzed at about the same time that the basic chemical makeup of DNA became known, in the 1940s. They were known to differ from DNA in three respects: the sugar in the molecule was ribose, instead of deoxyribose (hence the name); the molecule, instead of being double-stranded, was singlestranded (although there are some exceptions to this rule, notably in some viruses composed of RNA); and the thymine was replaced by a very similar base, uracil (Fig. 3-7). Several forms of RNA of different molecular weight (relative molecular mass) were known to exist in the cytoplasm and the nucleus. However, they appeared to be stable and, accordingly, not likely to fulfill the role of a messenger molecule that had to vary in length (and thus in molecular mass)
to reflect the complexity of the messages encoded in the DNA. The molecule that was finally identified as a messenger RNA, or mRNA, was difficult to discover because it constitutes only a small proportion of the total RNA (2% to 5%) and because of its relatively short life span. The DNA code is transcribed into mRNA with the help of specific enzymes, transcriptases (Fig. 3-8A). The transcription, which occurs in the nucleus on a single strand of DNA, follows the principles of nucleotide binding, as described for DNA replication, except that in RNA, thymine is replaced by a similar molecule, uracil (Fig. 3-8B). As will be set forth in the following section, each molecule of mRNA corresponds to one specific sequence of DNA nucleotides, encoding the formation of a single protein molecule, hence a gene. Because the size of the genes varies substantially, the mRNAs also vary in length, thus in molecular mass, corresponding to the length of the polypeptide chain to be produced in the cytoplasm. The identification of and, subsequently, the in vitro synthesis of mRNA proved to be critical in the further analysis of the genetic code and in subsequent work on analysis of the genetic activity of identifiable fragments of DNA. For a recent review of this topic, see articles by Cook (1999) and Klug (2001).
to reflect the complexity of the messages encoded in the DNA. The molecule that was finally identified as a messenger RNA, or mRNA, was difficult to discover because it constitutes only a small proportion of the total RNA (2% to 5%) and because of its relatively short life span. The DNA code is transcribed into mRNA with the help of specific enzymes, transcriptases (Fig. 3-8A). The transcription, which occurs in the nucleus on a single strand of DNA, follows the principles of nucleotide binding, as described for DNA replication, except that in RNA, thymine is replaced by a similar molecule, uracil (Fig. 3-8B). As will be set forth in the following section, each molecule of mRNA corresponds to one specific sequence of DNA nucleotides, encoding the formation of a single protein molecule, hence a gene. Because the size of the genes varies substantially, the mRNAs also vary in length, thus in molecular mass, corresponding to the length of the polypeptide chain to be produced in the cytoplasm. The identification of and, subsequently, the in vitro synthesis of mRNA proved to be critical in the further analysis of the genetic code and in subsequent work on analysis of the genetic activity of identifiable fragments of DNA. For a recent review of this topic, see articles by Cook (1999) and Klug (2001).
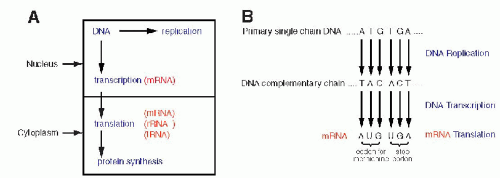 Figure 3-8 A. A diagrammatic representation of the principal nuclear and cytoplasmic events in protein formation. B. DNA replication, transcription, and translation for the amino acid methionine and for the stop codons, indicating the beginning and the end of protein synthesis. Note the replacement of thymine (T) by uracil (U) in mRNA. It is evident that the process could be reversed; by unraveling the composition of a protein and its amino acids, it is possible to deduce the mRNA condons, thereby the DNA code for this protein. |
Reannealing
In experimental in vitro systems, the bonds between the two chains of DNA can be broken by treatment with alkali, acids, or heat. Still, the affinity of the two molecules is such that once the cause of the strand separation is removed, the two chains will again come together, an event known as reannealing. These properties of the double-stranded DNA became of major importance in gene analysis and molecular engineering.
MOLECULAR TRAFFIC BETWEEN THE NUCLEUS AND THE CYTOPLASM
Although it has been known for many years that the nucleus is provided with gaps in its membrane, known as the nuclear pores (see Chap. 2 and Fig. 2-26), the precise function of the nuclear pores was unknown. Within recent years, some light has been shed on the makeup of the nuclear pores and on the mechanisms of transport between the nucleus and the cytoplasm. The nuclear pores are composed of complex molecules of protein that interact with DNA (Blobel, 1985; Gerace et al, 1978; Davies and Blobel, 1986). Further, specific molecules have been identified that assist in the export of mRNA and tRNA from the nucleus into the cytoplasm and import of proteins from the cytoplasm into the nucleus across the nuclear pores. Proteins, known as importins and exportins have now been identified as essential to the traffic between the nucleus and the cytoplasm. The interested readers are referred to a summary article by Pennisi (1998) and the bibliography listed.
The Genetic Code
The unraveling of the structure of DNA and its mechanism of replication was but a first step in understanding the mechanism
of cell function. The subsequent step required deciphering the message contained in the structure. Since neither the sugar molecule nor the phosphate molecule had any specificity, the message had to be contained in the sequence of the nucleotide bases (i.e., A,G,T, and C), as was suggested by Watson and Crick shortly after the fundamental discovery of the structure of the DNA. It was subsequently shown that the DNA code is limited to the formation of proteins from the 20 essential amino acids. The specific sequences of nucleotides that code for amino acids could be defined only after the pure form of the intermediate RNA molecules could be synthesized.
of cell function. The subsequent step required deciphering the message contained in the structure. Since neither the sugar molecule nor the phosphate molecule had any specificity, the message had to be contained in the sequence of the nucleotide bases (i.e., A,G,T, and C), as was suggested by Watson and Crick shortly after the fundamental discovery of the structure of the DNA. It was subsequently shown that the DNA code is limited to the formation of proteins from the 20 essential amino acids. The specific sequences of nucleotides that code for amino acids could be defined only after the pure form of the intermediate RNA molecules could be synthesized.
By a series of ingenious and deceptively simple experiments, it was shown that different clusters of three nucleotides coded for each of the 20 amino acids, the primary components of all proteins. A sequence of three nucleotides, encoding a single amino acid, is known as a codon (Fig 3-9B). A series of codons, corresponding to a single, defined polypeptide chain or protein, constitutes a gene. As discussed in the foregoing, the code inscribed in the DNA molecule is transcribed into mRNA, which carries the message into the cytoplasm of the cell wherein protein formation takes place (see Fig. 3-8A). The code, therefore, was initially defined, not as a sequence of nucleotides in the DNA, but as it was transcribed into RNA. Because there are four nucleotides in the RNA molecule (A,G,C, and U, substituting for T), and three are required to code for an amino acid, there are 4 X 4 X 4 or 64 possible combinations. These combinations could be established by using synthetic RNA. Thus, the identity of the triplets of nucleotides, each constituting a codon, could be precisely established (see Fig. 3-9). It may be noted that only one amino acid, methionine, is coded by a unique sequence, AUG (adenine, uracil, guanine). It was subsequently proven that the codon for methionine initiated the synthesis of a sequence of amino acids constituting a protein. In other words, every protein synthesis starts with a molecule of methionine, although this amino acid can be removed later from the final product. All other amino acids are encoded by two or more different codons. There are also three nucleotide sequences that are interpreted as termination or “stop” codons. The stop codons signal the end of the synthesis of a protein chain.
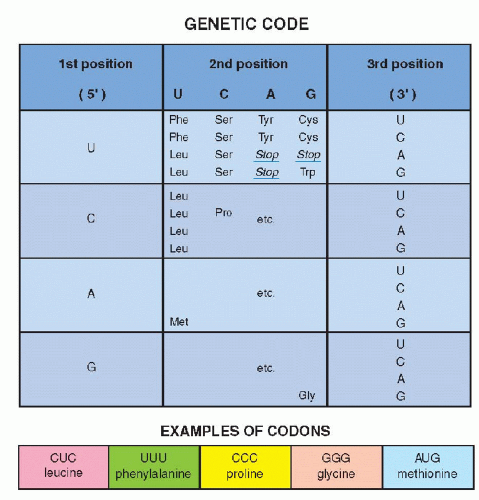 Figure 3-9 Examples of codons for several amino acids using the first, second, and third position of the mRNA nucleotides, uracil (U), cytosine (C), adenine (A), and guanine (G). It may be noted that 19 amino acids have multiple codes (for example, tyrosine [Tyr] is coded by UAU and UAC). There is but one code for methionine (Met), namely AUG, indicating the beginning of a protein. There are several stop codons, indicating the end of protein synthesis (see Fig. 3-8B). |
Once the RNA code was established, it became very simple to identify corresponding nucleotide sequences on the DNA by simply substituting U(racil) by T(hymine). This reciprocity between DNA and RNA base sequences was also subsequently utilized in further molecular biologic investigations (see Fig. 3-8B).
MECHANISMS OF PROTEIN SYNTHESIS OR mRNA TRANSLATION
The unraveling of the genetic code and the unique role of proteins still did not clarify the precise mechanisms of the synthesis of proteins, often composed of thousands of amino acids. It is now known that protein formation,
or translation of the message encoded in mRNA, takes place in the cytoplasm of the cell and requires two more types of RNA. One of these is ribosomal RNA (rRNA), which accounts for most of the RNA in the cell and is the principal component of ribosomes. These granulelike organelles are each made up of one small and one larger spherical structure separated by a groove, thus somewhat resembling a Russian doll (see Fig. 2-17). The third type of RNA is the transfer RNAs (tRNA), which function as carriers of the 20 specific amino acids that are floating freely in the cytoplasm of the cell. For a recent review of this topic, see the article by Cech (2000).
or translation of the message encoded in mRNA, takes place in the cytoplasm of the cell and requires two more types of RNA. One of these is ribosomal RNA (rRNA), which accounts for most of the RNA in the cell and is the principal component of ribosomes. These granulelike organelles are each made up of one small and one larger spherical structure separated by a groove, thus somewhat resembling a Russian doll (see Fig. 2-17). The third type of RNA is the transfer RNAs (tRNA), which function as carriers of the 20 specific amino acids that are floating freely in the cytoplasm of the cell. For a recent review of this topic, see the article by Cech (2000).
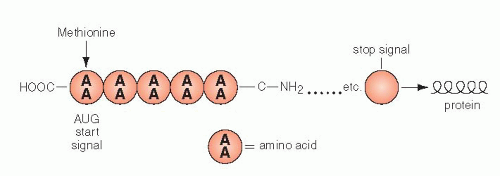
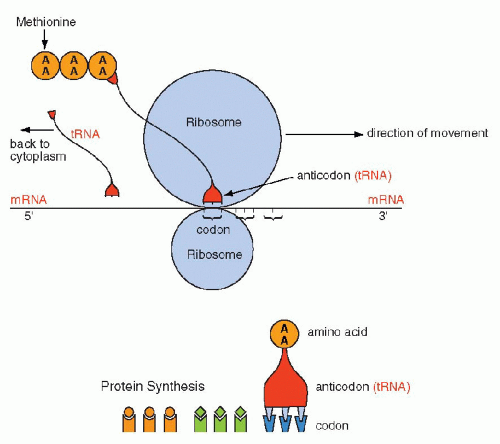 Figure 3-10 Schematic representation of protein formation. mRNA glides along a groove separating the two components of the ribosome in the 5′ to 3′ direction. Each codon is matched by an “anticodon,” carried by transfer RNA (tRNA), that one-by-one brings the amino acids encoded in mRNA to form a chain of amino acids or a protein. The protein synthesis begins with methionine and stops with a stop codon. Once the tRNA has delivered its amino acid, it is returned to the cytoplasm to start the cycle again. AA = amino acid. |
The synthesis of proteins occurs in the following manner: mRNA, carrying the message for the structure of a single protein, enters the cytoplasm, where it is captured by the ribosomes. The synthesis is initiated by the codon for methionine. The mRNA slides along the ribosomal groove, and the sequential codons are translated one by one into specific amino acids that are brought to it by tRNA. Each molecule of tRNA with its specific anticodon sequences that correspond to the codons, carries one amino acid (Fig. 3-10). In translation, the same principles apply to the matching (pairing) of nucleotides and the direction of synthesis, from the 5′ to the 3′ end, as those discussed for DNA replication and transcription into mRNA. The amino acids attach to each other by their carboxy (COOH)—and amino (NH)—terminals and form a protein chain. The synthesis stops when a stop codon is reached and the protein is released into the cytoplasm where it can be modified before use or export (Fig. 3-11). The specific sequence of events in translation is currently under intense scientific scrutiny. It is generally assumed that inaccurate translation results in formation of a so-called nonsense protein that is apparently recognized as such and is either not further utilized or is destroyed.
UNIQUENESS OF PROTEINS AS CELL BUILDING BLOCKS AND BASIS OF PROTEOMICS
The deciphering of the genetic code led to one inescapable conclusion: The code operates only for amino acids, hence proteins, and not for any other structural or chemical cell components, such as fats or sugars. Therefore, proteins, including a broad array of enzymes, are the core of all other cell activities and direct the synthesis or metabolism of all other cell constituents. By a feedback mechanism, the synthesis and replication of the fundamental molecules of DNA or RNA are also dependent on the 20 amino acids that form the necessary enzymes. Proteins execute all events in the cell and, thus, may be considered the plenipotentiaries of the genetic messages encoded in DNA and transmitted by RNA. One must reflect on the extraordinary simplicity
of this arrangement and the hierarchical organization that governs all events in life.
of this arrangement and the hierarchical organization that governs all events in life.
The recognition of the unique role of proteins in health and disease has led to the recently developed techniques of proteomics. The purpose of proteomics is the identification of proteins that may be specific for a disease process, leading to development of specific drugs (Liotta and Petricoin, 2000; Banks et al, 2000). Micromethods have been developed that allow protein extraction and identification from small fragments of tissue (Liotta et al, 2001).
DEFINITION OF GENES
Once the mechanism of protein formation had been unraveled, it became important to know more about the form in which the message is carried in the DNA. Briefly, from a number of studies, initially with the fruit fly, Drosophila, then with the mold, Neurospora, it could be demonstrated that each protein, including each enzyme, had its own genetic determinant, called a gene. With the discovery of the structure of DNA and the genetic code, a gene is defined as a segment of DNA, carrying the message corresponding to one protein or, by implication, one enzyme. The significance of the precise reproduction of the genetic message became apparent in 1949, when Linus Pauling and his colleagues suggested that sickle cell anemia, characterized by a deformity of the shape of red blood cells, was a “molecular disease.” The molecular nature of the disease was established some years later by Ingram, who documented that sickling was due to the replacement of a single amino acid (hence, by implication, one codon in several hundred) in two of the four protein chains in hemoglobin. This replacement changes the configuration of the hemoglobin molecule in oxygen-poor environments, with resulting deformity of the normal spherical shape of red blood cells into curved and elongated structures that resemble “sickles.” More importantly still, sickle cell anemia behaves exactly according to the principles of heredity established by Mendel. If only one parent carries the gene, the offspring has a “sickle cell trait.” If both parents carry the gene, the offspring develops sickle cell anemia.
To carry the implications of these observations still further, if the genes are segments of nuclear DNA, then they should also be detectable on the metaphase chromosomes. With the development of specific genetic probes and the techniques of in situ hybridization, to be described below, the presence of normal and abnormal genes on chromosomes could be documented.
REGULATION OF GENE TRANSCRIPTION: REPRESSORS, PROMOTERS, AND ENHANCERS
Once the principles of the structure, replication, and transcription of DNA were established, it became important to learn more about the precise mechanisms of regulation of these events. If one considers that the length of the DNA chain in an Escherichia coli bacterium is about four million base pairs and that of higher animals in excess of 80 million base pairs, these molecules must contain thousands of genes. How these genes are transcribed and expressed became the next puzzle to be solved. Since it appeared that the fundamental mechanisms could be the same, or similar, in all living cells regardless of species, these studies were initially carried out on bacteria, which offered the advantage of very rapid growth under controlled conditions that could be modified according to the experimental needs.
The French investigators, Jacob and Monod, demonstrated that the functions of genes controlling the utilization of the sugar, lactose, by the bacterium E. coli, depended on a feedback mechanism. The activation or deactivation of this mechanism depended on the presence of lactose in the medium. It was shown that the transcription of the gene encoding an enzyme (β-galactosidase) that is necessary for the utilization of lactose, is regulated by an interplay between two DNA sequences, the repressor and the operator. The activation or deactivation of the repressor function is vested in the operator. The repressor function, which prevents the activation of the family of enzymes known as transcriptases, is abolished at the operator site by the presence of lactose. In the absence of lactose, the repressor gene is active and blocks the transcription at the operator site. Once the operator gene is derepressed by the lactose, the β-galactosidase gene is transcribed into the specific mRNA by the enzyme, RNA polymerase. The activity of the RNA polymerase is triggered by two sequences of bases located on the DNA molecule, one about 35 and the other about 10 bases ahead of the site of transcription, or upstream. These DNA sequences are known as promoters and they are recognized by RNA polymerase as a signal that the transcription may begin downstream, that is, at the first nucleotide of the DNA sequence (gene) to be transcribed (Fig. 3-12). The promoter is provided with specific, very short nucleotide sequences, or “boxes,” which regulate still further the transcription of DNA into mRNA (the discussion of boxes will be expanded below). The terms upstream and downstream have become incorporated into the language of molecular biology to indicate nucleotide sequences located on the DNA either before or after a specified gene or sequence of genes.
In the cytoplasm of the bacterium, the mRNA, which contains the sequences necessary for the transcription of the β-galactosidase, together with two other adjacent genes (providing additional enzymes necessary for utilization of lactose by the bacterium) is transcribed into the three enzymes. The name operon was given to a sequence of the three genes that are transcribed into a single mRNA molecule. Subsequently, similar regulatory mechanisms were observed for other genes on prokaryotic cells, confirming the general significance of these observations.
The search for similar mechanisms in eukaryotic cells began soon thereafter. An important difference in mRNA between prokaryotic and eukaryotic cells must be stressed: The mRNA of prokaryotes contains information for several proteins (an operon), whereas the mRNA of eukaryotic cells
encodes only one protein, an advantage in the manipulation of this molecule.
encodes only one protein, an advantage in the manipulation of this molecule.
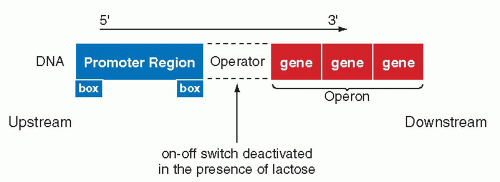 Figure 3-12 Regulation of the lac (lactose) gene expression in Escherichia coli. The transcription of the genes identified as operon, encoding the enzymes for utilization of the sugar lactose, may be blocked at a site named operator by a protein, the repressor, which is deactivated in the presence of lactose. The transcription of DNA into mRNA is initiated at a site known as the promoter region. The boxes indicate specific nucleotide sequences necessary in activation of RNA polymerase, the enzyme essential in transcription (see Fig. 2-14). |
Promoter sequences were also recognized in DNA of nucleated, eukaryotic cells. In such cells, two sequences of bases are known to occur: one of them is the so-called CAT box (a sequence of bases 5′-CCAAT-3′, occurring about 80 to 70 bases upstream, and the other, a TATA box (a sequence of 5′-TATAAA-3′), occurring about 30 to 25 bases upstream. The RNA polymerase activity begins at base 1, and it continues until the gene is transcribed. The end of the transcription is signaled by another box composed of AATAA sequence of bases (Fig. 3-13). At the beginning of the transcription, at its initial or 5′ site, the mRNA acquires a “cap” of methylguanidine residues, which presumably protects the newly formed molecule from being attacked by RNA-destroying enzymes (RNAses). At the conclusion of the transcription, the mRNA




Stay updated, free articles. Join our Telegram channel

Full access? Get Clinical Tree