Vector
Host
Insert size
Plasmid
E.coli
5–25 kb
λ phage
E.coli
35–45 kb
P1 phage
E.coli
70–100 kb
PACs
E.coli
100–300kb
BACs
E.coli
<300 kb
YACs
Saccharomyces cerevisae
200–2000kb
Human Artificial Chromosomes (HACs)
Cultured Human Cells
>2000kb
In order to determine the choice of vector for a particular cloning experiment, various factors need to be considered such as:
1.
Insert size: The insert size may vary for different types of vectors ranging from 5 to 25 kb for plasmid vectors to >2,000 kb for HACs.
2.
Vector size: The vector size range varies from 5 kb plasmid vectors to 6–10 megabases HAC high-capacity vectors.
3.
Restriction sites: The number of restriction sites found in vectors is highly variable. There may be a few restriction sites in small plasmid vectors but they may be increased by the insertion of multiple cloning sites in vectors.
4.
Copy number: Different cloning vectors are maintained at different copy numbers, dependent on the replicon of the plasmid. However, a high-copy number vector is desirable. The origin of replication determines the vector copy number, which could be in the range of 25–50 copies/cell if the expression vector is derived from the low-copy number plasmid pBR322, or between 150 and 200 copies/cell, if derived from the high-copy number plasmid pUC.
5.
Cloning efficiency: The ability to clone a DNA fragment inserted into a vector is known as the cloning efficiency of the vector.
6.
Ability to screen for inserts: For selection of recombinants, certain selectable markers should be present in vectors in order to distinguish them from non-recombinants.
7.
Types of downstream experiments required.
1.2 Vectors for Cloning Large Fragments of DNA
1.2.1 Cosmid Vectors
Cosmids are hybrids between a phage DNA molecule and a bacterial plasmid or are basically a plasmid that carries a cos site, the substrate for enzymes that package λ DNA molecule into phage coat proteins. The in vitro packaging reaction works not only with one genome but also with any DNA molecule that carries cos site separated by 37–52 kb of DNA. It also needs a selectable marker, such as ampicillin resistance gene, and a plasmid origin of replication, as cosmids lack all the λ genes, therefore do not produce plaques. Instead colonies are formed on selective media, just as with a plasmid vector. The loading capacity of cosmids varies depending on the size of the vector itself but usually lies around 40–45 kb—much more than a phage λ vector can accommodate. After packaging in vitro, the particle is used to infect suitable host. The recombinant cosmid DNA is injected into the cell where it circularizes like phage DNA but replicates as a normal plasmid without the expression of any phage functions. Transformed cells are selected on the basis of a vector drug resistance marker. The construct of a typical cosmid vector is shown in Fig. 1.1.
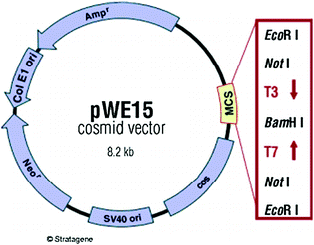

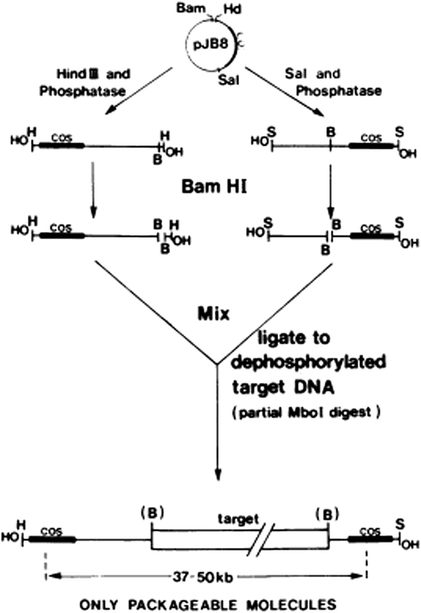
Fig. 1.1
Construct of a cosmid vector
Cosmids provide an efficient means of cloning large pieces of DNA. Because of their capacity to carry large fragments of DNA, cosmids are particularly attractive for constructing libraries of eukaryotic genome fragments. Partial digestion with a restriction endonuclease provided suitably large fragments. However, there is potential problem associated with use of partial digests in this way. This is due to the possibility of two or more genome fragments joining together in the ligation reaction, hence creating a clone containing fragments that were not initially adjacent in the genome. The problem can be overcome by the size fractionation and dephosphorylation of the foreign DNA fragments so as to prevent their ligation together. But this method is very sensitive to the exact ratio of target-to-vector DNAs because vector-to-vector ligation can occur. Such difficulties have been overcome in a cosmid-cloning procedure devised by Ish-Horowicz and Burke (1981). By appropriate treatment of the cosmid vector pJB8, left-hand and right-hand vector ends are purified which are incapable of self-ligation but which accept dephosphorylated foreign DNA. Thus, the method eliminates the need to size the foreign DNA fragments and prevents formation of clones containing short foreign DNA or multiple vector sequences. Figure 1.2 describes the cosmid-cloning procedure devised by Ish-Horowicz and Burke (1981).
Problems associated with lambda and cosmid cloning:
1.
Since repeats occur in eukaryotic DNA, rearrangements can occur via recombination of the repeats present on the DNA inserted into lambda or cosmid.
2.
Cosmids are difficult to maintain in a bacterial cell because they are somewhat unstable.
3.
Not easy to handle due to its very large size of approximately 50 kb.
1.2.2 Yeast Artificial Chromosomes
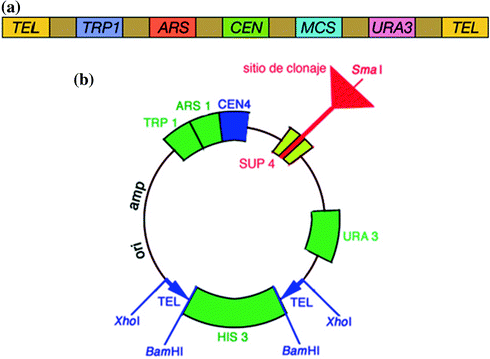
A YAC is a vector used to clone DNA fragments larger than 100 kb and up to 3,000 kb. YACs are useful for the physical mapping of complex genomes and for the cloning of large genes. First described in 1983 by Murray and Szostak, a YAC is an artificially constructed chromosome that contains a centromere (CEN), telomeres (TEL), and an autonomous replicating sequence (ARS) element which are required for replication and preservation of YAC in yeast cells. ARS elements are thought to act as replication origins. A YAC is built using an initial circular plasmid, which is typically broken into two linear molecules using restriction enzymes. DNA ligase is then used to ligate a sequence or gene of interest between the two linear molecules, forming a single large linear piece of DNA.
A plasmid-derived origin of replication (ori) and an antibiotic resistance gene allow the YAC vector to be amplified and selected for in E. coli. TRP1 and URA3 genes are included in the YAC vector to provide a selection system for identifying transformed yeast cells that include YAC by complementing recessive alleles trp1 and ura3 in yeast host cell. YAC vector cloning site for foreign DNA is located within the SUP4 gene. This gene compensates for a mutation in the yeast host cell that causes the accumulation of red pigment. The host cells are normally red, and those transformed with YAC only, will form colorless colonies. Cloning of a foreign DNA fragment into the YAC causes insertional inactivation, restoring the red color. Therefore, the colonies that contain the foreign DNA fragment are red.
1.2.2.1 Essential Components of YAC Vectors
1.
Large DNA (>100 kb) is ligated between two arms. Each arm ends with a yeast telomere so that the product can be stabilized in the yeast cell. Interestingly, larger YACs are more stable than shorter ones, which favors cloning of large stretches of DNA (Fig. 1.3a, b).