INTRODUCTION
Viruses are the smallest infectious agents (ranging from about 20 to 300 nm in diameter) and contain only one kind of nucleic acid (RNA or DNA) as their genome. The nucleic acid is encased in a protein shell, which may be surrounded by a lipid-containing membrane. The entire infectious unit is termed a virion. Viruses are parasites at the genetic level, replicating only in living cells and are inert in the extracellular environment. The viral nucleic acid contains information necessary to cause the infected host cell to synthesize virus-specific macromolecules required for the production of viral progeny. During the replicative cycle, numerous copies of viral nucleic acid and coat proteins are produced. The coat proteins assemble together to form the capsid, which encases and stabilizes the viral nucleic acid against the extracellular environment and facilitates the attachment and penetration by the virus upon contact with new susceptible cells. The virus infection may have little or no effect on the host cell or may result in cell damage or death.
The spectrum of viruses is rich in diversity. Viruses vary greatly in structure, genome organization and expression, and strategies of replication and transmission. The host range for a given virus may be broad or extremely limited. Viruses are known to infect unicellular organisms, such as mycoplasmas, bacteria, and algae, and all higher plants and animals. General effects of viral infection on the host are considered in Chapter 30.
Much information on virus–host relationships has been obtained from studies on bacteriophages, the viruses that attack bacteria. This subject is discussed in Chapter 7. Properties of individual viruses are discussed in Chapters 31–44.
TERMS AND DEFINITIONS IN VIROLOGY
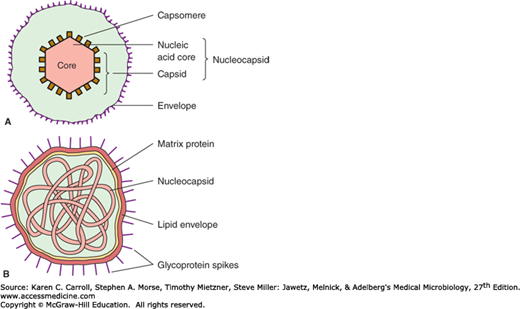
Schematic diagrams of viruses with icosahedral and helical symmetry are shown in Figure 29-1. Indicated viral components are described below.
Capsid: The protein shell, or coat, that encloses the nucleic acid genome.
Capsomeres: Morphologic units seen in the electron microscope on the surface of icosahedral virus particles. Capsomeres represent clusters of polypeptides, but the morphologic units do not necessarily correspond to the chemically defined structural units.
Defective virus: A virus particle that is functionally deficient in some aspect of replication.
Envelope: A lipid-containing membrane that surrounds some virus particles. It is acquired during viral maturation by a budding process through a cellular membrane (see Figure 29-3). Virus-encoded glycoproteins are exposed on the surface of the envelope. These projections are called peplomers.
Nucleocapsid: The protein–nucleic acid complex representing the packaged form of the viral genome. The term is commonly used in cases in which the nucleocapsid is a substructure of a more complex virus particle.
Structural units: The basic protein building blocks of the coat. They are usually a collection of more than one nonidentical protein subunit. The structural unit is often referred to as a protomer.
Subunit: A single folded viral polypeptide chain.
Virion: The complete virus particle. In some instances (eg, papillomaviruses, picornaviruses), the virion is identical with the nucleocapsid. In more complex virions (herpesviruses, orthomyxoviruses), this includes the nucleocapsid plus a surrounding envelope. This structure, the virion, serves to transfer the viral nucleic acid from one cell to another.
EVOLUTIONARY ORIGIN OF VIRUSES
The origin of viruses is not known. There are profound differences among the DNA viruses, the RNA viruses, and viruses that use both DNA and RNA as their genetic material during different stages of their life cycle. It is possible that different types of agents are of different origins. Two theories of viral origin can be summarized as follows:
Viruses may be derived from DNA or RNA nucleic acid components of host cells that became able to replicate autonomously and evolve independently. They resemble genes that have acquired the capacity to exist independently of the cell. Some viral sequences are related to portions of cellular genes encoding protein functional domains. It seems likely that at least some viruses evolved in this fashion.
Viruses may be degenerate forms of intracellular parasites. There is no evidence that viruses evolved from bacteria, although other obligately intracellular organisms (eg, rickettsiae and chlamydiae) presumably did so. However, poxviruses are so large and complex that they might represent evolutionary products of some cellular ancestor.
CLASSIFICATION OF VIRUSES
The following properties have been used as a basis for the classification of viruses. The amount of information available in each category is not the same for all viruses. Genome sequencing is now often performed early in virus identification, and comparisons with databases provide detailed information on the viral classification, predicted protein composition, and taxonomic relatedness to other viruses.
Virion morphology, including size, shape, type of symmetry, presence or absence of peplomers, and presence or absence of membranes.
Virus genome properties, including type of nucleic acid (DNA or RNA), size of the genome, strandedness (single or double), whether linear or circular, sense (positive, negative, ambisense), segments (number, size), nucleotide sequence, percent GC content, and presence of special features (repetitive elements, isomerization, 5′-terminal cap, 5′-terminal covalently linked protein, 3′-terminal poly(A) tract).
Genome organization and replication, including gene order, number and position of open reading frames, strategy of replication (patterns of transcription, translation), and cellular sites (accumulation of proteins, virion assembly, virion release).
Virus protein properties, including number, size, amino acid sequence, modifications (glycosylation, phosphorylation, myristoylation), and functional activities of structural and nonstructural proteins (transcriptase, reverse transcriptase, neuraminidase, fusion activities).
Antigenic properties, particularly reactions to various antisera.
Physicochemical properties of the virion, including molecular mass, buoyant density, pH stability, thermal stability, and susceptibility to physical and chemical agents, especially solubilizing agents and detergents.
Biologic properties, including natural host range, mode of transmission, vector relationships, pathogenicity, tissue tropisms, and pathology.
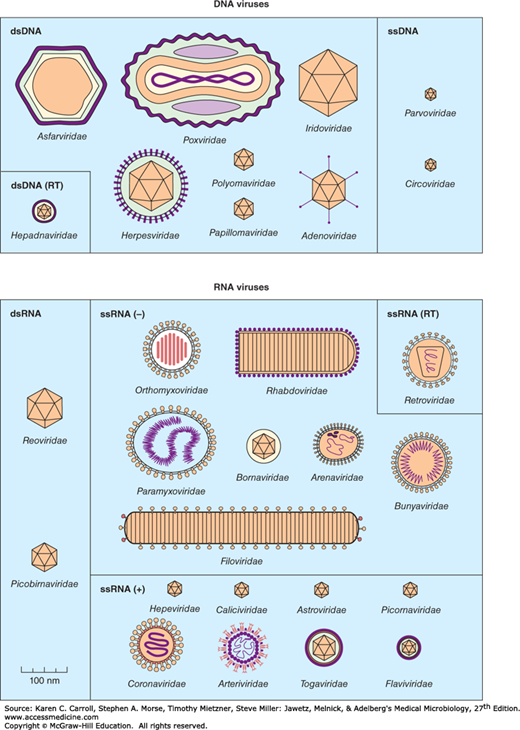
A system has been established in which viruses are separated into major groupings—called families—on the basis of virion morphology, genome structure, and strategies of replication. Virus family names have the suffix -viridae. Table 29-1 sets forth a convenient scheme used for classification. Diagrams of animal virus families are shown in Figure 29-2.
Figure 29-2
Shapes and relative sizes of animal viruses of families that infect vertebrates. In some diagrams, certain internal structures of the particles are represented. Only those families that include human pathogens are listed in Table 29-1 and described in the text. (Reproduced with permission from van Regenmortel MHV, Fauquet CM, Bishop DHL, et al [editors]: Virus Taxonomy: Classification and Nomenclature of Viruses. Seventh Report of the International Committee on Taxonomy of Viruses. Academic Press, 2000.)
| Nucleic Acid Core | Capsid Symmetry | Virion: Enveloped or Naked | Ether Sensitivity | Number of Capsomeres | Virus Particle Size (nm)a | Size of Nucleic Acid in Virion (kb/kbp) | Physical Type of Nucleic Acidb | Virus Family |
|---|---|---|---|---|---|---|---|---|
| DNA | Icosahedral | Naked | Resistant | 32 | 18–26 | 5.6 | ss | Parvoviridae |
| 30 | 2.0–3.9 | ss circular | Anelloviridae | |||||
| 72 | 45 | 5 | ds circular | Polyomaviridae | ||||
| 72 | 55 | 8 | ds circular | Papillomaviridae | ||||
| 252 | 70–90 | 26–45 | ds | Adenoviridae | ||||
| Enveloped | Sensitive | 180 | 40–48 | 3.2 | ds circularc | Hepadnaviridae | ||
| 162 | 150–200 | 125–240 | ds | Herpesviridae | ||||
| Complex | Complex coats | Resistantd | 230 × 400 | 130–375 | ds | Poxviridae | ||
| RNA | Icosahedral | Naked | Resistant | 32 | 28–30 | 7.2–8.4 | ss | Picornaviridae |
| 28–30 | 6.4–7.4 | ss | Astroviridae | |||||
| 32 | 27–40 | 7.4–8.3 | ss | Caliciviridae | ||||
| 27–34 | 7.2 | ss | Hepeviridae | |||||
| 35–40 | 4 | ds segmented | Picobirnaviridae | |||||
| 60–80 | 16–27 | ds segmented | Reoviridae | |||||
| Enveloped | Sensitive | 42 | 50–70 | 9.7–11.8 | ss | Togaviridae | ||
| Unknown or complex | Enveloped | Sensitive | 40–60 | 9.5–12.5 | ss segmented | Flaviviridae | ||
| 50–300 | 10–14 | Arenaviridae | ||||||
| 120–160 | 27–32 | ss | Coronaviridae | |||||
| 80–110 | 7–11e | ss diploid | Retroviridae | |||||
| Helical | Enveloped | Sensitive | 80–120 | 10–13.6 | ss segmented | Orthomyxoviridae | ||
| 80–120 | 11–21 | ss segmented | Bunyaviridae | |||||
| 80–125 | 8.5–10.5 | ss | Bornaviridae | |||||
| 75 × 180 | 13–16 | ss | Rhabdoviridae | |||||
| 150–300 | 16–20 | ss | Paramyxoviridae | |||||
| 80 × 1000f | 19.1 | ss | Filoviridae |
Within each family, subdivisions called genera are usually based on biological, genomic, physicochemical, or serologic differences. Criteria used to define genera vary from family to family. Genus names carry the suffix -virus. In several families (Herpesviridae, Paramyxoviridae, Parvoviridae, Poxviridae, Reoviridae, Retroviridae), a larger grouping called subfamilies has been defined, reflecting the complexity of relationships among member viruses. Virus orders may be used to group virus families that share common characteristics. For example, order Mononegavirales encompasses the Bornaviridae, Filoviridae, Paramyxoviridae, and Rhabdoviridae families. As of 2013, the International Committee on Taxonomy of Viruses had organized more than 2500 animal and plant virus species into 103 families and 455 genera.
Properties of the major families of animal viruses that contain members important in human disease are summarized in Table 29-1. They are discussed briefly below in the order shown in Table 29-1 and are considered in greater detail in the chapters that follow.
Parvoviruses (from Latin parvus meaning small) are very small viruses with a particle size of about 18–26 nm. The particles have cubic symmetry, with 32 capsomeres, but they have no envelope. The genome is linear, single-stranded DNA, averaging 5 kb in size. Replication occurs only in actively dividing cells; capsid assembly takes place in the nucleus of the infected cell. Human parvovirus B19 replicates in immature erythroid cells and causes several adverse consequences, including aplastic crisis, fifth disease, and fetal death. (See Chapter 31.)
Anelloviruses (from Latin anello meaning ring) are small (~30 nm in diameter), icosahedral virions that lack an envelope. The viral genome is negative sense, circular, single-stranded DNA, 2–4 kb in size. Anelloviruses include the torque teno viruses, and are globally distributed in the human population and many animal species. No specific disease associations have been proven.
Polyomaviruses are small (45 nm), nonenveloped, heat-stable, solubilization-resistant viruses exhibiting cubic symmetry, with 72 capsomeres. The name derives from Greek poly– (many) and –oma (tumor) and refers to the ability of some of these viruses to produce tumors in infected hosts. The genome is circular, double-stranded DNA, about 5 kb in size. These agents have a slow growth cycle, stimulate cell DNA synthesis, and replicate within the nucleus. The most well-known human polyomaviruses are JC virus, the causative agent of progressive multifocal leukoencephalopathy; BK virus, associated with nephropathy in transplant recipients; and Merkel cell virus, found associated with the majority of Merkel cell skin carcinomas. SV40, a primate virus, can also infect humans. Most animal species harbor chronic infections with one or more polyomaviruses. (See Chapter 43.)
Papillomaviruses are similar to polyomaviruses in some respects but with a larger genome (8 kb) and particle size (55–60 nm). The name refers to Latin papilla (nipple) and Greek –oma (tumor) and describes wart-like lesions produced by these viral infections. Certain types of human papillomaviruses are causative agents of genital cancers in humans. Papillomaviruses are very host and tissue specific and cannot be grown in cultured cells in vitro. Many animal species carry papillomaviruses. (See Chapter 43.)
Adenoviruses (from Latin adenos meaning gland) are medium-sized (70–90 nm), nonenveloped viruses exhibiting cubic symmetry, with fiber spikes protruding from capsomers that aid in host attachment. The genome is linear, double-stranded DNA, 26–48 kb in size. Replication occurs in the nucleus. Complex splicing patterns produce mRNAs. At least 57 types infect humans, especially in mucous membranes, and some types can persist in lymphoid tissue. Adenoviruses can cause acute respiratory diseases, conjunctivitis, and gastroenteritis. Some human adenoviruses can induce tumors in newborn hamsters. Many serotypes infect animals. (See Chapters 32 and 43.)
Hepadnaviruses (from Latin hepa meaning liver) are small (40–48 nm), enveloped viruses containing circular, partially double-stranded DNA molecules that are about 3.2 kbp in size. Replication involves repair of the single-stranded gap in the DNA, transcription of RNA, and reverse transcription of the RNA to make genomic DNA. The virus consists of a 27-nm icosahedral nucleocapsid core within a closely adherent envelope that contains lipid and the viral surface antigen. The surface protein is characteristically overproduced during replication of the virus, which takes place in the liver, and is shed into the bloodstream. Hepadnaviruses such as Hepatitis B virus can cause acute and chronic hepatitis; persistent infections are associated with a high risk of developing liver cancer. Viral types are known that infect mammals and ducks. (See Chapter 35.)
Herpesviruses are family of large viruses 150–200 nm in diameter. The name refers to Latin herpes (creep), describing the spreading nature of skin lesions caused by these viruses. The nucleocapsid is 100 nm in diameter, with cubic symmetry and 162 capsomeres, surrounded by a lipid-containing envelope. The genome is linear, double-stranded DNA, 120–240 kb in size. Latent infections may last for the life span of the host, usually in ganglial or lymphoblastoid cells. Human herpesviruses include herpes simplex types 1 and 2 (oral and genital lesions), varicella-zoster virus (chickenpox and shingles), cytomegalovirus, Epstein-Barr virus (infectious mononucleosis and association with human neoplasms), human herpesviruses 6 and 7 (T cell lymphotropic), and human herpesvirus 8 (associated with Kaposi sarcoma). Other herpesviruses occur in many animals. (See Chapters 33 and 43.)
Poxviruses are large brick-shaped or ovoid viruses 220–450 nm long × 140–260 nm wide × 140–260 nm thick. The particle structure is complex, with a lipid-containing envelope. The name derives from Anglo-Saxon pokkes meaning pouch, referring to their characteristic vesicular skin lesions. The genome is linear, covalently closed, double-stranded DNA, 130–375 kb in size. Poxvirus particles contain about 100 proteins, including many with enzymatic activities, such as a DNA-dependent RNA polymerase. Replication occurs entirely within the cell cytoplasm. Some are pathogenic for humans (smallpox, vaccinia, molluscum contagiosum); others that are pathogenic for animals can infect humans (cowpox, monkeypox). (See Chapter 34.)
Picornaviruses are small (28–30 nm), ether-resistant viruses exhibiting cubic symmetry. The RNA genome is single stranded and positive sense (ie, it can serve as an mRNA) and is 7.2–8.4 kb in size. The groups infecting humans are enteroviruses (polioviruses, coxsackieviruses, echoviruses, and rhinoviruses [more than 100 serotypes causing common colds]) and hepatovirus (hepatitis A). Rhinoviruses are acid labile and have a high density; other enteroviruses are acid stable and have a lower density. Picornaviruses infecting animals include foot-and-mouth disease of cattle and encephalomyocarditis of rodents. (See Chapter 36.)
Astroviruses are similar in size to picornaviruses (28–30 nm), but particles display a distinctive star-shaped outline on their surfaces. The genome is linear, positive-sense, single-stranded RNA, 6.8–7.0 kb in size. These agents may be associated with gastroenteritis in humans and animals. (See Chapter 37.)
Caliciviruses are similar to picornaviruses but slightly larger (27–40 nm). The particles appear to have cup-shaped depressions on their surfaces. The genome is single-stranded, positive-sense RNA, 7.3–8.3 kb in size; the virion has no envelope. Important human pathogens are the noroviruses (eg, Norwalk virus), the cause of epidemic acute gastroenteritis. Other agents infect cats and sea lions as well as primates. (See Chapter 37.)
Hepeviruses are similar to caliciviruses. The particles are small (32–34 nm) and ether resistant. The genome is single-stranded, positive-sense RNA, 7.2 kb in size. It lacks a genome-linked protein (VPg). Human hepatitis E virus belongs to this group. (See Chapter 35.)
Picobirnaviruses are small (35–40 nm) nonenveloped viruses with icosahedral structure. The genome is linear, double-stranded, segmented (bipartite) RNA (two segments), totaling about 4 kb.
Reoviruses are medium-sized (60–80 nm), ether-resistant, nonenveloped viruses having icosahedral symmetry. Particles have two or three protein shells with channels extending from the surface to the core; short spikes extend from the virion surface. The genome is linear, double-stranded, segmented RNA (10–12 segments), totaling 18–30 kbp in size. Individual RNA segments range in size from 200 to 3000 bp. Replication occurs in the cytoplasm; genome segment reassortment occurs readily. Reoviruses of humans include rotaviruses, which have a distinctive wheel-shaped appearance and cause gastroenteritis. Antigenically similar reoviruses infect many animals. The genus Coltivirus includes Colorado tick fever virus of humans. (See Chapter 37.)
Arboviruses and rodent-borne viruses are ecologic groupings (not a virus family) of viruses with diverse physical and chemical properties. The arboviruses (there are more than 350 of them) have a complex cycle involving arthropods as vectors that transmit the viruses to vertebrate hosts by their bite. Viral replication does not seem to harm the infected arthropod. Arboviruses infect humans, mammals, birds, and reptiles, and use mosquitoes and ticks as vectors. Human pathogens include dengue, yellow fever, West Nile fever, and encephalitis viruses. Rodent-borne viruses establish persistent infections in rodents and are transmitted without an arthropod vector. Human diseases include hantavirus infections and Lassa fever. The viruses in these ecologic groupings belong to several virus families, including arenaviridae, bunyaviridae, flaviviridae, reoviridae, rhabdoviridae, and togaviridae. (See Chapter 38.)
Many arboviruses that are major human pathogens, called alphaviruses—as well as rubella virus—belong to this group. They have a lipid-containing envelope and are ether sensitive, and their genome is single-stranded, positive-sense RNA, 9.7–11.8 kb in size. The enveloped virion measures 65–70 nm. The virus particles mature by budding from host cell membranes. An example is eastern equine encephalitis virus. Rubella virus has no arthropod vector. (See Chapters 38 and 40.)
Flaviviruses are enveloped viruses, 40–60 nm in diameter, containing single-stranded, positive-sense RNA. Genome sizes vary from 9.5 to 12 kb. Mature virions accumulate within cisternae of the endoplasmic reticulum. This group of arboviruses includes yellow fever virus and dengue viruses. Most members are transmitted by blood-sucking arthropods. Hepatitis C virus has no known vector. (See Chapters 35 and 38.)
Arenaviruses are pleomorphic, enveloped viruses ranging in size from 60 to 300 nm (mean, 110–130 nm). The genome is segmented, circular, single-stranded RNA that is negative sense and ambisense, 10–14 kb in total size. Replication occurs in the cytoplasm with assembly via budding on the plasma membrane. The virions incorporate host cell ribosomes during maturation, which gives the particles a “sandy” appearance. Most members of this family are unique to tropical America (ie, the Tacaribe complex). All arenaviruses pathogenic for humans cause chronic infections in rodents. Lassa fever virus of Africa is one example. These viruses require maximum containment conditions in the laboratory. (See Chapter 38.)
Coronaviruses are enveloped 120- to 160-nm particles containing an unsegmented genome of positive-sense, single-stranded RNA, 27–32 kb in size. Coronaviruses resemble orthomyxoviruses but have petal-shaped surface projections arranged in a fringe, similar to a solar corona. Coronavirus nucleocapsids develop in the cytoplasm and mature by budding into cytoplasmic vesicles. These viruses have narrow host ranges. Most human coronaviruses cause mild acute upper respiratory tract illnesses—“colds”—but a new coronaviruses causes severe acute respiratory syndrome (SARS) and Middle East respiratory syndrome (MERS). Toroviruses, which cause gastroenteritis, form a distinct genus. Coronaviruses of animals readily establish persistent infections and include mouse hepatitis virus and avian infectious bronchitis virus. (See Chapter 41.)
Retroviruses are spherical, enveloped viruses (80–110 nm in diameter) whose genome contains two copies of linear, positive-sense, single-stranded RNA. Each monomer RNA is 7–11 kb in size. Particles contain a helical nucleocapsid within an icosahedral capsid. Replication is unique; the virion contains a reverse transcriptase enzyme that produces a DNA copy of the RNA genome. This DNA becomes circularized and integrated into host chromosomal DNA. The virus is then replicated from the integrated “provirus” DNA copy. Virion assembly occurs by budding on plasma membranes. Hosts remain chronically infected. Retroviruses are widely distributed; there are also endogenous proviruses resulting from ancient infections of germ cells transmitted as inherited genes in most species. Leukemia and sarcoma viruses of animals and humans (see Chapter 43), foamy viruses of primates, and lentiviruses (human immunodeficiency viruses; visna of sheep) (see Chapters 42 and 44) are included in this group. Retroviruses cause acquired immunodeficiency syndrome (AIDS) (see Chapter 44) and make possible the identification of cellular oncogenes (see Chapter 43).
Orthomyxoviruses are medium-sized, 80- to 120-nm enveloped viruses exhibiting helical symmetry. Particles are either round or filamentous, with surface projections that contain hemagglutinin or neuraminidase activity. The genome is linear, segmented, negative-sense, single-stranded RNA, totaling 10–13.6 kb in size. Segments range from 890 to 2350 nucleotides each. The virus matures by budding at the cell membrane. Orthomyxoviruses include influenza viruses that infect humans or animals. The segmented nature of the viral genome permits ready genetic reassortment when two influenza viruses infect the same cell, presumably fostering the high rate of natural variation among influenza viruses. Viral reassortment and transmission from other species is thought to explain the emergence of new human pandemic strains of influenza A viruses. (See Chapter 39.)
Bunyaviruses are spherical or pleomorphic, 80- to 120-nm enveloped particles. The genome is made up of a triple-segmented, single-stranded, negative-sense or ambisense RNA, 11–19 kb in overall size. Virion particles contain three circular, helically symmetric nucleocapsids about 2.5 nm in diameter and 200–3000 nm in length. Replication occurs in the cytoplasm, and an envelope is acquired by budding into the Golgi. The majority of these viruses are transmitted to vertebrates by arthropods (arboviruses). Hantaviruses are transmitted not by arthropods but by persistently infected rodents via aerosols of contaminated excreta. They cause hemorrhagic fevers and nephropathy as well as a severe pulmonary syndrome. (See Chapter 38.)
Bornaviruses are enveloped, spherical (70–130 nm) viruses. The genome is linear, single-stranded, nonsegmented, negative-sense RNA, 8.5–10.5 kb in size. Unique among nonsegmented, negative-sense RNA viruses, replication and transcription of the viral genome occur in the nucleus. Borna disease virus is neurotropic in animals; a postulated association with neuropsychiatric disorders of humans is unproven. (See Chapter 42.)
Rhabdoviruses are enveloped virions resembling a bullet, flat at one end and round at the other, measuring about 75 × 180 nm. The envelope has 10-nm spikes. The genome is linear, single-stranded, nonsegmented, negative-sense RNA, 11–15 kb in size. Particles are formed by budding from the cell membrane. Viruses have broad host ranges. Rabies virus is a member of this group. (See Chapter 42.)
Paramyxoviruses are similar to but larger (150–300 nm) than orthomyxoviruses. Particles are pleomorphic. The internal nucleocapsid measures 13–18 nm, and the linear, single-stranded, nonsegmented, negative-sense RNA is 16–20 kb in size. Both the nucleocapsid and the hemagglutinin are formed in the cytoplasm. Those infecting humans include mumps, measles, parainfluenza, metapneumo, and respiratory syncytial viruses. These viruses have narrow host ranges. In contrast to influenza viruses, paramyxoviruses are genetically stable. (See Chapter 40.)
Filoviruses are enveloped, pleomorphic viruses that may appear very long and threadlike. They typically are 80 nm wide and about 1000 nm long. The envelope contains large peplomers. The genome is linear, negative-sense, single-stranded RNA, 18–19 kb in size. Marburg and Ebola viruses cause severe hemorrhagic fever in Africa. These viruses require maximum containment conditions (Biosafety Level 4) for handling. (See Chapter 38.)
Novel viruses are being discovered with increasing frequency; most belong to existing families but rarely agents are not classifiable. Some of these are associated with human disease, while many affect other species. (See Chapter 48.)
Viroids are small infectious agents that cause diseases of plants. Viroids are agents that do not fit the definition of classic viruses. They are nucleic acid molecules without a protein coat. Plant viroids are single-stranded, covalently closed circular RNA molecules consisting of about 360 nucleotides and with a highly base-paired rodlike structure. Viroids replicate by an entirely novel mechanism. Viroid RNA does not encode any protein products; the devastating plant diseases induced by viroids occur by an unknown mechanism. Hepatitis D virus in humans has properties similar to viroids.
Prions are infectious particles composed solely of protein with no detectable nucleic acid. They are highly resistant to inactivation by heat, formaldehyde, and ultraviolet light that inactivate viruses. The infectious prion protein is misfolded and able to change the conformation of the native prion protein which is encoded by a single cellular gene. Prion diseases, called “transmissible spongiform encephalopathies,” include scrapie in sheep, mad cow disease in cattle, and kuru and Creutzfeldt-Jakob disease in humans. (See Chapter 42.)
PRINCIPLES OF VIRUS STRUCTURE
Viruses come in many shapes and sizes. Structural information is necessary for virus classification and for establishing structure–function relationships of viral proteins. The particular structural features of each virus family are determined by the functions of the virion: morphogenesis and release from infected cells; transmission to new hosts; and attachment, penetration, and uncoating in newly infected cells. Knowledge of virus structure furthers our understanding of the mechanisms of certain processes such as the interaction of virus particles with cell surface receptors and neutralizing antibodies. It may lead also to the rational design of antiviral drugs capable of blocking viral attachment, uncoating, or assembly in susceptible cells.
Electron microscopy, cryoelectron microscopy, and x-ray diffraction techniques have made it possible to resolve fine differences in the basic morphology of viruses. The study of viral symmetry by standard electron microscopy requires the use of heavy metal stains (eg, potassium phosphotungstate) to emphasize surface structure. The heavy metal adsorbs to virus particles and brings out the surface structure of viruses by virtue of “negative staining.” The typical level of resolution is 3–4 nm. (The size of a DNA double helix is 2 nm.) However, conventional methods of sample preparation often cause distortions and changes in particle morphology. Cryoelectron microscopy uses virus samples quickly frozen in vitreous ice; fine structural features are preserved, and the use of negative stains is avoided. Three-dimensional structural information can be obtained by the use of computer image processing procedures. Examples of image reconstructions of virus particles are shown in following chapters. (See Chapters 32 and 37.)
X-ray crystallography can provide atomic resolution information, generally at a level of 0.2–0.3 nm. The specimen must be crystalline, and this has only been achieved with small, nonenveloped viruses. However, it is possible to obtain high-resolution structural data on well-defined substructures prepared from the more complex viruses.
Stay updated, free articles. Join our Telegram channel

Full access? Get Clinical Tree