Plate type
Advantage
Drawback
Proposed application
96-Well plate
Large number of cells per well, area ~30 mm2
Low number of wells
Validation screen with preselected low number of targets
384-Well plate
Medium number of wells
–
Genome-wide screen
Cell arrays
Full assay on a single plate. No need for plate handling. Cell seeding and antibody staining simultaneously on whole plate
Relatively small area (~0.13 mm) per spot, ~50–300 cells
Genome-wide screen with expensive reagents or rare patient cells
3.1 Preparation of the Source siRNA Transfection Solution for Solid-Phase Transfection
1.
Prepare an siRNA stock solution by dissolving lyophilized siRNAs with MilliQ water to a final concentration of 30 μM.
2.
Transfer 3 μl of OptiMEM, containing 0.4 M sucrose, to each well of a 384-well low-volume plate.
3.
Add 3.5 μl of Lipofectamin 2000 to each well of the 384-well low-volume plate and mix thoroughly.
4.
Add 5 μl of the respective siRNA stock solution to each well of the 384-well low-volume plate and mix thoroughly.
5.
Incubate for 20 min at room temperature.
6.
Add 7.25 μl of a 0.2 % gelatin solution containing 1 × 10−² % fibronectin to each well of the 384-well low-volume plate and mix thoroughly.
3.2 Preparation of “Ready to Transfect” Plates
1.
Option 1: Preparation of 96-well plates “ready to transfect”:
Dilute 18.75 μl of the source siRNA transfection solution with 468.75 μl MilliQ water and dispense 25 μl per well of a 96-well plate.
Option 2: Preparation of 384-well plates “ready to transfect”:
Dilute 18.75 μl of the source siRNA transfection solution with 187.5 μl MilliQ water and dispense 5 μl per well of a 384-well plate.
Option 3: Preparation of high-density cell arrays “ready to transfect”:
The siRNA transfection solution is not further diluted and printed directly onto the cell array by a full contact printer (see Note 2 ). No further steps are needed!
3.3 Comparison of Different Screening Microscopes
Various types of microscopes can be used for screening approaches. Crucial factors for the choice of the right system are the imaging speed and the quality of the readout. First we want to give a comparison between three different fluorescence-based screening systems, a wide-field (Olympus, IX81), a spinning disc confocal (PerkinElmer, Opera), and a confocal (Leica, SP5) microscope. On each system, the same screen was performed. A 96-well plate (ibidi) was used on which a layout with 12 wells (see Fig. 1) was repeated 3 times, one repetition for each microscope screen. In each well, multiple subpositions were acquired until the total recorded area per well covered about 1,240 × 930 μm2. At each subposition a z-stack with 24 steps (z-step width 2 μm) was recorded for three different color channels. The imaging parameters of the different microscopes are compared in Table 2.
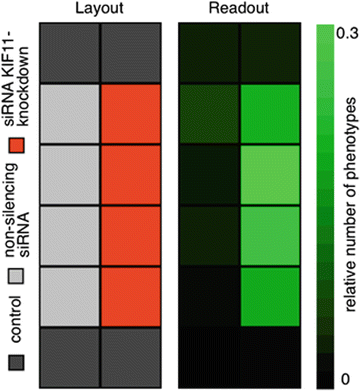

Fig. 1
Layout of the plate and readout example. Four wells are used as controls where no siRNA was applied (dark gray, top and bottom row), in four wells a non-silencing siRNA was applied (light gray, left column), and in four wells siRNA for Kif11 knockdown was transfected (red, right column), which resulted in prometaphase arrest of the cells. The relative number of cells in prometaphase is counted and the relative amount compared to all imaged cells is determined
Table 2
Imaging parameter for three different microscope systems
Imaging parameter
Stay updated, free articles. Join our Telegram channel
Full access? Get Clinical Tree
 Get Clinical Tree app for offline access
Get Clinical Tree app for offline access

|
|---|