CHAPTER 23 Degradation of Cellular Components*
An individual cell can live for weeks, months, years, or even the entire lifetime of the organism, but the cell’s constituent proteins, lipids, and RNA turn over continuously. This process of molecular degradation and replacement serves three functions. Constitutive turnover is a “housekeeping” function that ensures regular replacement of older molecules with newly synthesized ones or that removes misfolded, mislocalized, or otherwise damaged molecules so that they do not hinder the function of native molecules. Regulated or induced turnover results in rapid degradation of specific target molecules and functions in signal transduction, regulation of the cell cycle, and remodeling of cells and tissues during development. Finally, macroautophagy, a more global mechanism for degradation of cellular proteins or lipids, can be triggered under conditions of starvation, when the cell perceives a shortage of specific raw materials, such as amino acids. This chapter focuses primarily on the mechanisms that govern protein degradation and turnover, as these are the best studied; lipid turnover is also discussed. Chapter 16 covers RNA turnover.
Characteristics of Constitutive Protein Turnover
The turnover of cellular constituents at steady state arises from a careful balance of the rates of synthesis and degradation. Turnover can be massive. For example, approximately 40% of total cellular protein in rat liver is degraded every day. The rates of transcription and translation determine the rate of synthesis of each protein. Once synthesized, each type of protein turns over at a characteristic rate. Degradation is random, so older molecules are not selected over younger ones. The time course of degradation of the population of any specific protein follows a single exponential—strong evidence that chance, rather than aging, determines which copies of the protein are degraded (see Fig. 4-1 for first-order reactions). The rate is usually expressed as a half-life, the time for half of the molecules to be degraded.
The intrinsic rate of degradation of a given protein is determined by many factors, including its size, overall charge, thermal instability, flexibility, hydrophobicity, folding, and assembly with other protein subunits (if it is multimeric). Long-lived proteins have half-lives measured in days, while short-lived proteins have half-lives of hours or minutes. Small, basic proteins tend to have longer half-lives than do large, acidic proteins, and key enzymes of metabolic pathways often have very short half-lives. A protein may have specific sequences or structural motifs that are recognized by the proteolytic machinery. Phosphorylation marks some proteins for destruction. The rate at which a protein is degraded can be altered either by increasing the activity of its degradative pathway or by exposure or creation of “degradation motifs” on the protein to initiate destruction.
Degradation in Lysosomes
Lysosomes, the major digestive organelles, contain at least 60 distinct hydrolytic enzymes, including proteases, lipases, phospholipases, glycosidases, and nucleases. Lysosomal hydrolases are tagged in the Golgi apparatus with mannose-6-phosphate groups on their N-linked oligosaccharides. Mannose-6-phosphate re-ceptors in the trans-Golgi network bind lysosomal hydrolases for diversion to endosomes and lysosomes (see Chapter 22). Most lysosomal hydrolases are synthesized as inactive precursors and are activated by proteolysis on arrival in lysosomes. The low pH of lysosomes, maintained by the vacuolar adenosine triphosphatase (ATPase) proton pump (see Fig. 8-5C), is essential for efficient degradation. Most lysosomal enzymes have maximal hydrolytic activity at pH 4 to 5 rather than at the cytoplasmic pH of 6.5 to 7.0. Moreover, the low pH helps to denature many proteins, increasing their susceptibility to degradation. It is important to note that the hydrolases themselves are more resistant than are most macromolecules to the harsh environment. Lysosomal enzymes degrade proteins, lipids, and nucleic acids to fragments that are small enough to be transported either actively or passively across the lysosomal membrane to the cytoplasm, where they are reused to synthesize new macromolecules.
Lysosomes degrade substrates that originate both outside and inside the cell. Extracellular substrates taken into the cell by endocytosis are delivered to lysosomes via the endocytic pathway (see Chapter 22). Lysosomes also degrade cellular constituents, accounting for 50% to 70% of cellular protein turnover. Substrates are delivered to lysosomes by endocytosis and by direct translocation from the cytoplasm. Degradation of intracellular substrates is generally termed autophagy, which occurs by three distinct mechanisms called crinophagy, macroautophagy, and microautophagy, which are described later.
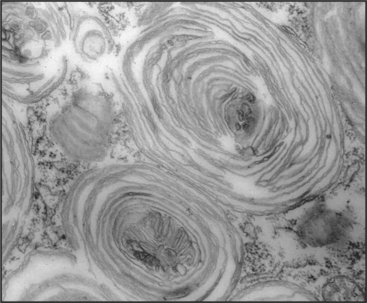
The essential role of lysosomes as the primary site for constitutive degradation is revealed by the more than 30 distinct human lysosomal storage diseases (Appendix 23-1). Patients with these diseases lack the function of one or more lysosomal hydrolases. Consequently, undigested material accumulates in lysosomes and causes them to swell (Fig. 23-1), ultimately killing the cell.
Delivery to Lysosomes via the Endocytic Pathway
Lysosomal degradation of plasma membrane proteins internalized by endocytosis plays an important role in remodeling the plasma membrane in response to cell stimuli. For example, the half-life of the receptor for the epidermal growth factor (EGF) is normally approximately 10 hours. However, when circulating EGF binds, the activated receptor is more efficiently internalized and degraded in lysosomes with a half-time of less than 1 hour. This downregulates the biological response (see Fig. 27-6).
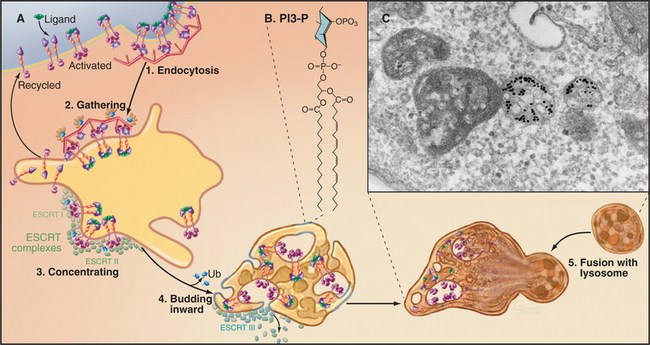
The degradation of lipids and membrane proteins in lysosomes poses a topologic problem, since fusion of lysosomes with other membrane compartments would simply merge the two membranes. This problem is solved by segregating the components to be degraded into regions of membrane that bud into endosomes, forming the intraluminal vesicles or tubules of multivesicular bodies (see Fig. 22-16). Fusion of a multivesicular body with a lysosome delivers the intraluminal vesicles to the lumen of the lysosome for digestion (Fig. 23-2). Resident lysosomal membrane proteins remain in the limiting membrane.
The formation of multivesicular bodies starts with a sorting process in endosomes (see Chapter 22). Proteins that are destined for degradation are tagged with single ubiquitin molecules, a process that is distinct from the polyubiquitination reaction required for targeting to proteasomes (see later). These monoubiquitinated proteins are gathered together in endosomes by ubiquitin-binding proteins that are localized there by interaction with phosphatidylinositol-3-phosphate (PI3-P), formed by PI3-kinase (see Fig. 26-7). A sequence of three multiprotein ESCRT complexes (endosomal com-plex required for transport [Fig. 22-16]) then further concentrates the monoubiquitinated proteins on regions of membrane that bud into the vesicle interior by an unknown mechanism. Components of the ESCRT complexes initially bind monoubiquitin but later recruit enzymes that remove and recycle the ubiquitin. Interestingly, budding of a number of viruses, including HIV-1, the virus that causes AIDS, resembles the process of multivesicular body formation. In fact, HIV hijacks two of the ESCRT complexes for this purpose.
Autophagy
Microautophagy and macroautophagy both describe the consumption of cytoplasmic constituents within lysosomes. Microautophagy occurs as a by-product of the formation of multivesicular bodies (Fig. 23-2; also see Fig. 22-16). Small volumes of cytoplasm are captured in the intraluminal vesicles and tubules that invaginate within endosomal or lysosomal membranes. The cytoplasmic components are degraded as the vesicles are consumed. However, studies in yeast suggest the exist-ence of alternative pathways for the selective packaging and delivery of cytosolic proteins to lysosomes. When starved for glucose, Saccharomyces cerevisiae expresses several cytosolic enzymes and membrane transporters that are required to process more complex sugars. When glucose becomes available, the yeast switches metabolic pathways and degrades the enzymes it no longer needs. Transporters in the plasma membrane are internalized and delivered to the vacuole (the yeast lysosome equivalent) through the formation of multivesicular bodies and microautophagy. Unneeded cytoplasmic enzymes are selectively packaged into small vesicles that deliver their contents to the vacuole by fusion. Genetic studies of yeast are unraveling the mechanisms of these rapidly induced and selective degradation pathways.
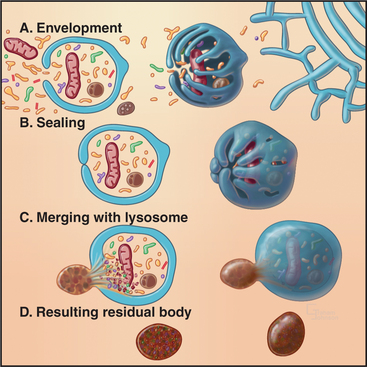
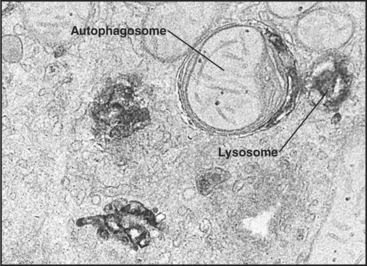
Macroautophagy involves the engulfment of large regions of cytoplasm—that might include glycogen granules, ribosomes, and organelles, such as mitochondria and peroxisomes—into an autophagic vacuole. Autophagic vacuoles begin to form when a flattened membrane cisterna encircles a region of cytosol and closes into a vesicle with two membranes (Fig. 23-3). The origin of the smooth membrane cisternae is uncertain and could be either the smooth endoplasmic reticulum (ER), the trans-Golgi apparatus, or the endosomal system. The trans-Golgi apparatus has the advantage of having targeting information for fusion with lysosomes. Fusion of a nascent autophagic vacuole with late endosomes and lysosomes forms an autolysosome with acid hydrolases in the lumen to degrade the contents (Figs. 23-3 and 23-4). The end stage of an autolysosome is typically a residual body with a dense core of undegraded material. The process of formation and degradation of autophagic vacuoles in the liver requires less than 15 minutes.
Because large volumes of cytoplasm and entire organelles are destroyed, macroautophagy must be regulated precisely and directly. The intracellular signal that triggers macroautophagy is thought to be tied to the intracellular levels of particular amino acids, which are, in turn, related to the extracellular concentrations of these amino acids. Amino acids are potent inhibitors of autophagy. Circulating peptide hormones also regulate autophagy by binding receptors and activating signaling cascades that involve protein phosphorylation. For example, starvation increases the circulating levels of the hormone glucagon, which stimulates autophagy in liver cells. Feeding produces the opposite reaction by increasing the levels of insulin, which reduce auto-phagy. Diurnal feeding rhythms cause the numbers of autophagic vacuoles to vary along with the fluctuation of essential amino acids in the blood.
Budding yeast use autophagy as a cell survival pathway during starvation, but autophagy can kill some cells. For example, autophagy can lead to death when a cell receives a lethal insult under conditions in which apoptotic pathways (see Chapter 46) are not functional. Whether autophagy is a physiological cell death pathway in addition to apoptosis remains controversial. Autophagy has also been implicated in the developmental programs of a number of higher organisms and in the destruction of intracellular protein aggregates.