Fig. 9.1.
Barcode scanning at gross station equipped with barcoding scanners for material tracking.
Historical Perspective
1960–2000
♦
Interdisciplinary field at the cross section of biology, medicine, computer science, statistics and mathematics, and information technology
♦
Traditional definition of biomedical informatics includes computational techniques to solve biological problems:
Therefore, biomedical informatics is not a new discipline, and the earliest examples include studies on protein sequences
♦
With the advent of sequencing technologies, computational methods to study genomic sequences were developed
♦
One such method is the BLAST sequence alignment algorithm
♦
DNA sequences can be used for the study of comparative genomics, evolutionary genomics, biological diversity, protein prediction, biological annotation, and the findings of mutations in cancer and other diseases
♦
Complex software tools and databases are being developed for these studies
♦
The National Center for Biotechnology Information (NCBI) was established in 1988 (http://www.ncbi.nlm.nih.gov/):
NCBI provides/creates computational tools for biomedical research such as GenBank for sequence data
♦
Because of large-scale genome sequencing projects including the human and other genomes, there has been exponential growth in the volume of data deposited into GenBank
♦
Development of high-throughput experimental methods, such as expression chips, allows for simultaneous measurement of thousands of molecular events
2000 Onwards
♦
A number of genomic processes can be studied using high-throughput “omic” technologies :
These include DNA sequence, methylation, copy number changes, single-nucleotide polymorphisms (SNPs), microRNA levels, mRNA expression levels, long noncoding RNAs, transcriptional complexity, chromatin conformation, and protein-DNA binding
Next-generation sequencing (NGS) technologies are replacing chip-based technologies for these studies. NGS techniques include RNA Seq for expression profiling, ChiP Seq for protein-DNA binding, exome and whole-genome sequencing (WXS, WGS) for DNA single-nucleotide polymorphism (SNP), structural variation, and copy number detection
♦
Proteomic methods using profiling of thousands of serum or tissue markers can be quantitated simultaneously using mass spectrometry
♦
Tissue microarrays (TMAs) , where multiple biospecimen cores are placed on glass slides, can be used to study protein expression in multiple samples simultaneously
♦
NGS data are considered big data and can be in the terabyte range for a single experiment and require sophisticated algorithms and computational infrastructure
♦
There are a number of national and international biomedical informatics projects to create databases, tools, and profile genomic changes in diseases such as cancer :
Large database efforts such as NCBI, Ensembl (http://www.ensembl.org/index.html), and UCSC genome databases (http://genome.ucsc.edu/) are free online resources for highly annotated genomic data for vertebrate and other eukaryotes
Examples of large-scale consortia projects included the Cancer Genome Atlas (TCGA) (cancergenome.nih.gov) whose aim is to characterize genomic changes in at least 500 cases of over 20 different cancer types
Nucleic Acids Research publishes an annual issue devoted to biomedical informatics databases
Laboratory Information System Overview
♦
The laboratory information system (LIS) provides an efficient method for managing information for enabling and disseminating diagnosis for the patients encountered within the surgical pathology practice
♦
There are many dedicated vendors offering a broad range of LIS solutions:
The anatomic pathology laboratory information system (APLIS) may exist as a separate stand-alone entity with interface to other hospital information systems or may be a module within a larger all-inclusive clinical laboratory or hospital-wide system
Both these solutions are applicable for a wide spectrum of surgical pathology practices including academic and private practices
♦
LIS functionality has been progressing significantly with added enhancements enabling the handling and distribution of pathology reports more easily:
There are three categorie s: basic functions that are to be expected in almost any system, advanced functionality, and future functionality
Tools
Imaging in Anatomic Pathology
♦
Imaging is a key component of an anatomic pathology practice along with other biomedical informatics tools:
It is an emerging technology that has the potential to modify and improve diagnostic methods in pathology
♦
Advancement in imaging technology is due to an overall improvement in computer technology, more rapid networking, and cheaper storage; soon we will arrive at a time when pathologists are able to manage images almost as easily and flexibly as they are able to handle text
♦
Imaging systems have the capability to provide provisions such as the permanent storage capability of static images and their incorporation in signout and reporting, including multiple cameras, network connectivity to an image server and the pathologists’ workstations, a storage device and database, and image capture and image display software
♦
These systems are largely used in gross image management and for microscopic single field documentation
♦
Imaging software is highly diverse and performs many functions:
Image processing software permits the adjustment of brightness, contrast, hue, and resolution, and that is very helpful to embellish the pathologic images for academic use (Fig. 9.2)
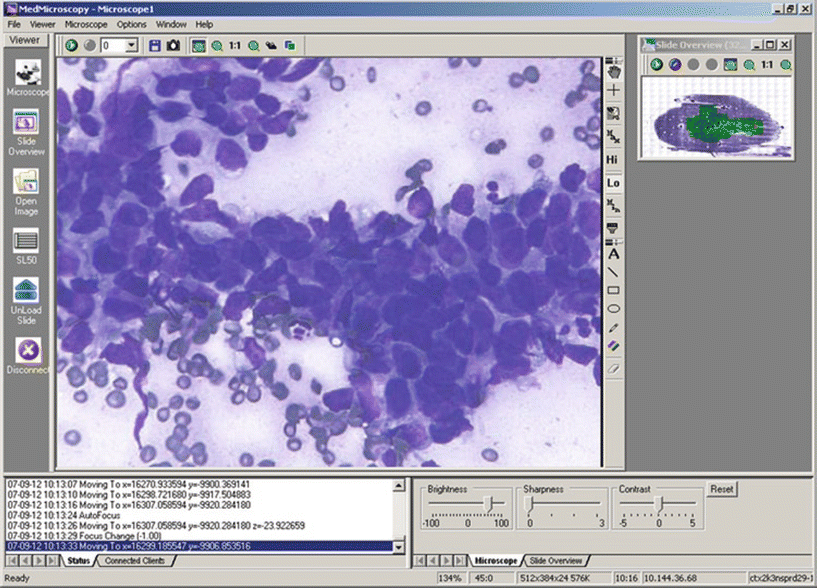
Fig. 9.2.
Whole slide image of fine-needle aspirate from a patient with adenocarcinoma, diagnosed by remote consultant.
♦
Image-embedded reports assist in patient care by providing detailed information with inclusion of graphs and diagrams:
These reports allow the clinicians to have better understanding of the pathology practice, augment more understanding between disciplines, and serve as a powerful marketing tool for expanding the boundaries of one’s practice
♦
Images are commonly used in many areas of daily pathology practice such as gross pathology of autopsy and surgical pathology specimens and for the microscopic documentation of normal and abnormal tissues (Fig. 9.3)

Fig. 9.3.
Biomedical informatics-driven workflow creates automation leading to increased efficiency and reduction in patient errors.
Telepathology
♦
Development in video microscopy and digital imaging capabilities enables light microscopic examination and diagnosis at remote sites
♦
General approaches adopted in telepathology are dynamic, static, and hybrid systems, each having its own advantages and disadvantages
♦
Dynamic systems are essentially remote-controlled microscopes that give the pathologist a live view of the distant microscopic image while allowing him or her to move the stage, focus, and change the magnification remotely:
The lucid advantage of dynamic systems is that they give the pathologist great flexibility in viewing the entire slide, to examine in detail any specific field at any power
Disadvantages of dynamic telepathology systems include the expensive and proprietary nature of the host and client stations and the great bandwidth needed to carry live, full-motion video
♦
Static (“store-and-forward”) telepathology requires that the referring pathologist captures a collection of still digital images for transmission to the consultant:
The major disadvantage is the loss of control experienced by the consulting telepathologist, who must rely on the referring pathologist to capture all of the necessary diagnostic fields to support an adequate examination
Static systems can be very flexible
Any system that captures images in standard file formats, when combined with any file transfer protocol program or adequate e-mail system, can be used to perform static telepathology
♦
Hybrid systems are developed by combining some of the better features of both static and dynamic systems:
Hybrid telepathology systems combine with remote-controlled high-resolution microscopy still image capture and retrieval
This approach requires a considerably lower bandwidth than purely dynamic systems while providing high-quality static images for diagnostic, reporting, and medical record-keeping purposes
♦
The systems (dynamic and hybrid) that provide better control to the consultant pathologist appear to present better results in terms of both increased diagnostic accuracy and lowered deferral rates
♦
Developments in telepathology have focused on three areas:
The first is the acceptance of open standards that will allow intersystem operability
The second area is the incorporation of telepathology into the LIS to facilitate image database reporting and billing capabilities
The third area is the development of whole slide imaging (WSI) for use in clinical practice
♦
Recently, guidelines for telepathology have been published by the American Telemedicine Association (ATA) (Pantanowitz et al. 2014):
The purpose of these guidelines is to assist users in pursuing a safe and sound process when practicing telepathology
Whole Slide Imaging
♦
In the late 1990s, pathologists began to experiment with systems that imaged and permanently stored the entire slide at high resolutions
♦
In 1997, a system in which a robotic microscope captured a large area of a slide, field by field, and a computer that “knitted” the individual fields together into a montage was developed
♦
This system had significant limitations; however, the most important one was the duration of prolonged time needed to capture a single extended field
♦
In late 1999, a prototype with a fully functional robot was developed
♦
It was based on traditional microscope optics, a strobe light connected to a precision stage, and a digital video camera
♦
Primary magnification of 20, a numerical aperture of 0.7, and square, 6.6 µm pixels; it had a spatial sampling period (pixel size/optical magnification – a measure of resolution) of 0.33 μm/pixel and could image a slide in 5–10 min depending on the size of the tissue section and the amount of image compression desired
♦
Image quality produced is generally of diagnostic quality, and with the viewing software, clinical annotations and clinical metadata presented with the image are made possible with potential resulting in a virtual microscope with all the clinical information needed to sign out the case (Fig. 9.4)

Fig. 9.4.
Automation and standardization of workflow.
♦
Today , a typical imaging robot can perform in batch mode (reading barcodes on slides) and can capture and compress an image of a slide with a 1.5 × 1.5 cm tissue section in approximately 6 min with spatial sampling periods between 0.3 and 0.5 μm/pixel
♦
Latest devices are implementing nontraditional optics, illumination, and sensors designed specifically for very high-speed image capture and should result in significant improvements in speed, throughput, and resolution in the months and years ahead, with different manufacturers eventually focusing on different aspects of the market
♦
Recently, the College of American Pathologists (CAP) have published guidelines for validation of WSI for clinical applications (Pantanowitz et al. 2013):
These guidelines, which are based on scientific evidence and expert opinion, will allow pathologists to now move closer toward actually using validated WSI technology in a safe manner to improve patient care
♦
Recent studies have established the utility of WSI in teaching and assessment:
The high-speed WSI robot industry is becoming highly diverse with a broad range of optics, detectors , slide handling devices, and software resulting in an increasing range of capabilities and costs (Fig. 9.5)
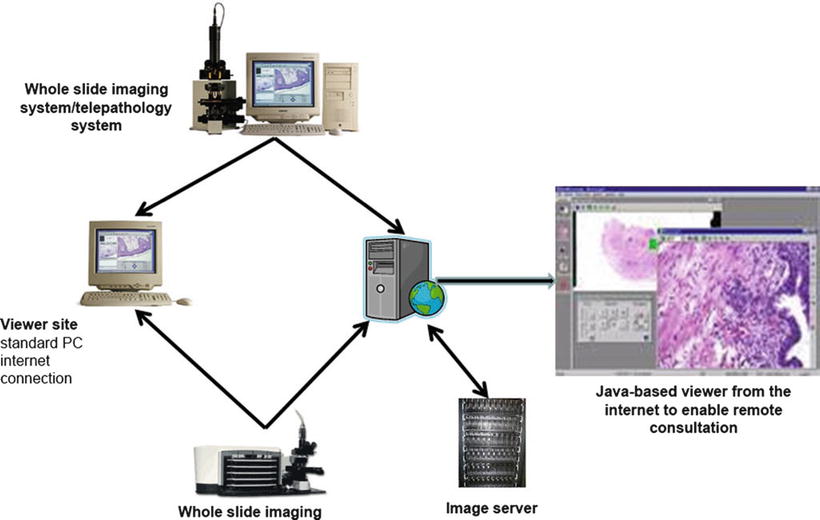
Fig. 9.5.
Imaging robots are networked to storage.
Structured Data Entry (Synoptic Reporting)
♦
Gross and microscopic examination of surgical specimens, particularly large resections, produces detailed information with implication for ongoing and future medical and oncology care :
A significant proportion of use of prognostic information about patient care is obtained from pathology reports
♦
Narrative descriptive reports have been used traditionally in surgical pathology to provide valuable information to the patients and their health-care teams:
This information allows great value in making treatment decisions such as adjuvant therapy, radiation, chemotherapy, prognosis, and outcomes
♦
Data capturing in the LIS is usually carried out by dictation followed by transcription:
Result in the creation of a few large text fields correlating with specific parts of the surgical pathology report (e.g., gross description, microscopic description, diagnosis)
The dictation is usually free form, with the pathologist speaking in continuous sentences, guided directly by the material at hand
Traditional narrative and descriptive reports in free text format have significant variability because different pathologists use a multitude of different reporting styles to describe their findings and variability results in pathology reports missing important data elements such as margins, lymphatic invasion, etc
♦
Synoptic reporting provides uniform standardized data elements in forms of checklists to ensure that pathologists make note of these findings in their reports:
Structured data entry takes a different approach and holds the promise of specific benefits. This model can be used to drive synoptic reports to support data entry:
•
The basic idea is that data is entered in many smaller specific text fields rather than in a few large ones
This mode of entry leads to supported data-querying capabilities, automated analysis, decision support, and predefined comment generation
LIS vendors are currently employing tools for synoptic reporting within their systems:
•
These systems have not reached their full-fledged potential, and there remains an area for further development
Synoptic reports provide a great degree of freedom to the pathologist to use structured data; for anything other than synoptic reporting, it is difficult because of placement in one large text field
The College of American Pathologists’ cancer protocols and checklists were created with the objective of improving the quality and uniformity of information in pathology reports:
♦
LIS does not maintain discrete data elements for synoptic data elements:
The pathology checklists provide unstructured text blocks, which are embedded in the pathology reports
The latter arrangement results in the presentation of pertinent pathology data in a cumbersome and difficult-to-access format
♦
Dictated with structured data, use a template that parses every single data element into its own predefined place in the database
♦
Synoptic report can either be placed at the end of the surgical pathology report or can replace the conventional free text-based final diagnosis section
♦
Structured collection of data could allow great versatile queries because every discrete data element (e.g., 1.8 cm) is directly linked to its inherent context
♦
Structured data entry offers significant benefits for reporting as well:
The template could easily define a synopsis of diagnostic findings
Stay updated, free articles. Join our Telegram channel

Full access? Get Clinical Tree


