8.1 INTRODUCTION TO ASSEMBLY AND EXIT OF VIRIONS FROM CELLS
In this chapter we deal with the final stages of the replication cycle: assembly of virions and their exit from the cell.
A virion has to be a stable structure as it has to survive in the environment as an infectious entity (though there is huge variation amongst viruses in the degree of stability, as discussed in Chapter 24). There is, however, also a requirement for a virion to become unstable during infection of a host cell so that the viral genome can be released. The virion must therefore have a built-in “switch” that can initiate the change from stability to instability when the appropriate environment is encountered. The “trigger” that activates the “switch” might be binding to a receptor and/or a change in the concentration of H+ or other ions.
Once threshold quantities of progeny virus genomes and structural proteins have accumulated in the infected cell, assembly of virions can commence. These components are assembled into nucleocapsids. If the virion also contains lipid then the assembly process also includes the acquisition of this component, either as an internal membrane or as an envelope.
8.2 NUCLEOCAPSID ASSEMBLY
8.2.1 Helical viruses
The assembly of virions and nucleocapsids of ssRNA viruses with helical symmetry (Section 3.4.1) involves coating copies of the genome with multiple copies of a protein, e.g. coat protein for tobacco mosaic virus, nucleoprotein for influenza viruses. Detailed studies of the assembly of tobacco mosaic virus have shown that discs of coat protein are formed first, and then these build up on the RNA to form the rod-shaped virion. The genomes of minus-strand RNA viruses become associated with additional proteins, such as RNA polymerases.
8.2.2 Icosahedral viruses
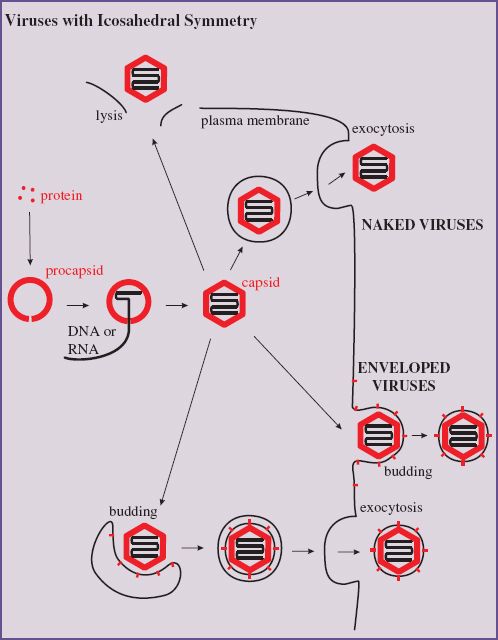
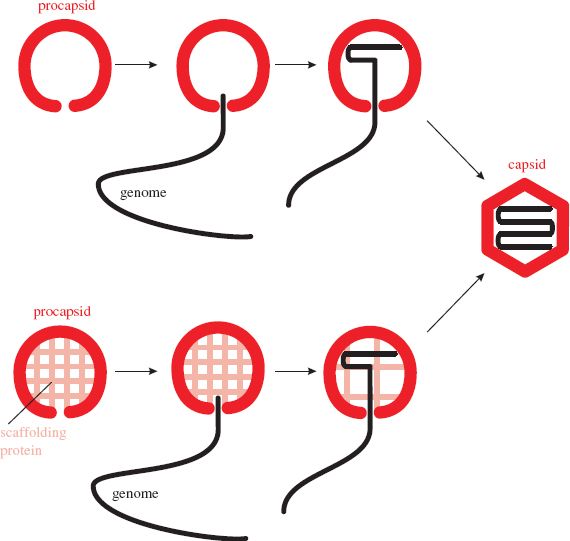
The assembly of virions and nucleocapsids of many viruses with icosahedral symmetry (Section 3.4.2) involves the construction of an empty protein shell, known as a procapsid, or a prohead in the case of a tailed bacteriophage. The procapsid is filled with a copy of the virus genome (Figure 8.1); during or after this process it may undergo modification to form the mature capsid. Modification of the procapsid may result in a change from a spherical to an icosahedral shape. For some viruses, including adenoviruses and picornaviruses, modification of the procapsid involves cleavage of one or more of the structural proteins.
Figure 8.1 Assembly of an icosahedral nucleocapsid. A protein shell (procapsid) is constructed. A copy of the virus genome enters the procapsid, which undergoes modifications to form the capsid. The procapsid of some viruses is built around scaffolding protein, which is removed as the genome is packaged.
The genome enters the procapsid through a channel located at a site that will become one of the twelve vertices of the icosahedron. Any enzymes involved in packaging the genome are located at this site. In the procapsids of herpesviruses and tailed phages the channel consists of a “portal” composed of one or more protein species (Section 3.4.2.d). The portal contains a powerful motor that moves the virus DNA into the procapsid.
8.2.3 Genome packaging
In the context of packaging virus genomes during virion assembly the question “How are virus genomes selected from all the cell and virus nucleic acids?” is an intriguing one. In a retrovirus-infected cell, for example, less than 1% of the RNA is virus genome. In fact, retroviruses do package a variety of cell RNAs, including tRNA, which plays a key role when the next host cell is infected (Chapter 17). Some other viruses also package cell nucleic acids; for example, tailed phages sometimes package host DNA.
For the most part, however, cell nucleic acids are not packaged. For some viruses it has been shown that this is achieved through the recognition by a virus protein of a specific virus genome sequence, known as a packaging signal. In single-stranded genomes the packaging signal is within a region of secondary structure. Most viruses with single-stranded genomes package either the plus-strand or the minus-strand, so the packaging signal must be present only in the strand to be packaged.
It is important that some copies of the virus genome are not packaged: those that are serving as templates for synthesis of DNA, RNA, or protein. The packaging signal must be masked in these copies of the genome.
Virus genomes are packaged into small volumes. The dsDNA of large icosahedral viruses, such as herpesviruses and the tailed phages, is packed so tightly that the pressure within the capsid is about ten times greater than the pressure inside a champagne bottle.
The tight packaging of viral genomes means that repulsion between the negative charges on their phosphate groups must be overcome. This may be achieved by packaging basic proteins, which are positively charged, along with the genome. Some ssRNA viruses (e.g. rhabdoviruses, influenza viruses, and retroviruses) coat their genomes with basic protein, while some dsDNA viruses (e.g. adenoviruses and baculoviruses) package a basic protein in close association with the genome. All of these basic proteins are virus encoded, but papillomaviruses and polyomaviruses coat their DNA with cell histones, the basic proteins associated with the DNA in eukaryotic cells. Some viruses package other positively charged materials, such as polyamines and cations.
8.2.4 Assembly mechanisms
In the laboratory, virions can be disassembled into their component molecules. For some viruses infectious virions can be reassembled from the purified components (protein and nucleic acid), under appropriate conditions of pH and in the presence of certain ions. Virions that can self-assemble in this way are those with a relatively simple structure, composed of a nucleic acid and one or a small number of protein species. Viruses that can self-assemble in a test tube are assumed to undergo self-assembly in the infected cell. Examples are tobacco mosaic virus (helical symmetry) and the ssRNA phages (icosahedral symmetry). Self-assembly is economical because no additional genetic information is needed for the assembly process.
The virions of more complex viruses (e.g. herpes-viruses and the tailed phages) do not reassemble from their components in a test tube. The environment within the infected cell is required and the virions are constructed by a process of directed assembly.
Directed assembly of icosahedral viruses may involve proteins that are temporarily present while the virion is under construction, but are not present in the mature virion. These proteins are known as scaffolding proteins (Figure 8.1). Once their job is completed they are removed from the procapsid. Some are removed by proteolysis, while others are removed intact and can be recycled. Scaffolding proteins of the tailed phages play roles in determining the size and shape of the phage head.
Some viruses synthesize polyproteins from which the individual virus proteins are cleaved by proteases; many of the cleavages take place during the assembly process. Picornaviruses and retroviruses are two groups of viruses that utilize this strategy. The final cleavage of a picornavirus polyprotein does not occur until a copy of the genome enters the procapsid (Section 14.4.5), while the internal structures of the retrovirus virion are formed from polyprotein cleavage products late in the assembly process (Chapter 17).
8.3 FORMATION OF VIRION MEMBRANES
Enveloped virions acquire their membrane envelopes by one of two mechanisms; either they modify a host cell membrane and then nucleocapsids bud through it, or the virus directs synthesis of a new membrane, which forms around the nucleocapsids.
8.3.1 Budding through cell membranes
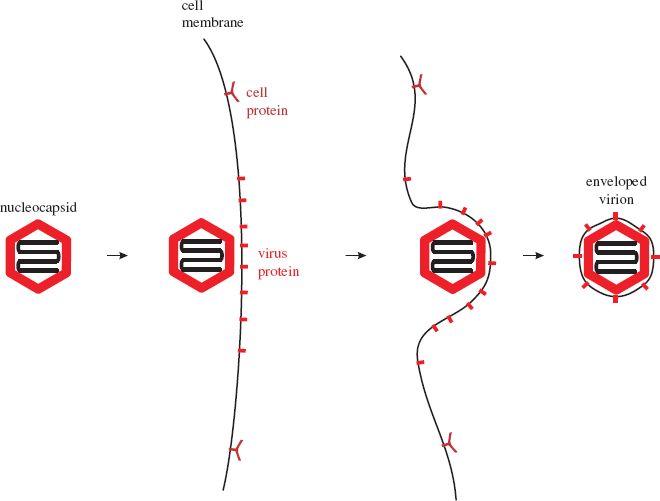
Most enveloped viruses acquire their envelopes by budding through a membrane of the host cell (Figure 8.2). For viruses with eukaryotic hosts this membrane is often the plasma membrane; the virions of most retroviruses and rhabdoviruses acquire their envelopes in this way. Membrane regions where budding will occur become modified by the insertion of one or more species of virus protein, the vast majority of which are glycoproteins (Section 6.4.3.a). Nucleocapsids accumulate adjacent to the regions of membrane containing virus protein.
Figure 8.2 Acquisition of a virion envelope by budding. Virus membrane proteins, usually glycoproteins, become incorporated into regions of a cell membrane, often the plasma membrane. A nucleocapsid buds through the membrane, which pinches off to form the virion envelope.
Stay updated, free articles. Join our Telegram channel

Full access? Get Clinical Tree


