Figure 17.3.1 Generation of a reporter construct to identify inhibitors of cell-to-cell signaling. (A) This panel represents the native promoter and gene that is regulated by CCS. (B)This panel demonstrates how the construct should be made to fuse a general CCS promoter to a reporter gene. The retention of an initiation codon from the endogenous gene ensures the continued regulation of the construct by the CCS promoter. (C). This panel demonstrates the fusion used in the specific example in the unit. The ler promoter, regulated by endogenous and exogenous signals, is fused to the gene encoding the β-galactosidase gene, lacZ.
General Growth Conditions
Since many CCS systems are also linked to growth phase, one must take particular note of the phase of growth in identifying inhibitory compounds. To be able to compare the day-to-day results, a standard growth protocol must be established. This protocol would usually start with an overnight growth, which is then diluted on the day of the experiment and allowed to grow to a predetermined level of growth as measured either by optical density or by colony-forming units (cfu). Additionally, the same medium would be used in the assays each day. The ideal strain to use in the identification of CCS inhibitors would lack the gene or genes responsible for the production of the signaling molecule. The assay conditions will include the addition of the signaling molecule at a close to saturating concentration, and the production of a significant endogenous signaling molecule may lead to difficulty in data interpretation. If the gene or genes responsible for the production of the signaling molecule are not known or cannot be determined, then the selection of a point in the growth curve becomes essential. One will select a point in the growth curve that contains enough cells to provide a robust signal in the output, but has relatively little endogenous signaling molecule. Usually, this point will be in early logarithmic growth. For the EHEC example, we have not identified the genes responsible for the production of the AI-3 molecule to date and, as such, we use a point early in the logarithmic growth.
Choice of media is also a key feature. Growth in a medium that does not result in the proper CCS-dependent phenotype will make the identification of CCS inhibitors difficult. Chemically defined media are most desirable as there are often fewer components to interfere with signal development. Additionally, the treatment of the potential inhibitory compounds should be the same. Each compound should be suspended in an identical diluent. The addition of various diluents in the assay introduces a variable that cannot be well controlled for in a high-throughput manner.
Overall, establishing a standard growth protocol will lead to fewer false-positive and false-negative results based on controllable assay conditions.
In Vitro Identification of Inhibitors of Cell-Cell Communication Compounds
General methodology
Once an acceptable reporter system and growth conditions have been established, one can start to screen potential inhibitor compounds. Knowledge of the signaling compound at this point in the identification of inhibitors can provide a checkpoint for the selection of the compounds to screen. If the signaling molecule is chemically defined, it may be possible to chemically synthesize potential inhibitors rather than perform an unbiased screening of a large library of compounds. Knowledge of the chemical structure of the signaling molecule will allow identification of the active structural units, and if combined with data regarding the binding pocket on the receptor, very potent inhibitors can be designed. This biased screening method may reduce the number of compounds that will have to be screened, but it also prevents the discovery of novel compounds that may inhibit the CCS system. The unbiased screening of libraries of compounds works well when there is little or no knowledge of the signaling molecule. The most common libraries that are now being utilized are the small molecule libraries. These consist of a large number (>100,000) of small-molecular-weight compounds that have been synthesized either as the library or for another purpose. By using an unbiased approach in the screening of the library, one may identify compounds that provide clues to the structure of the endogenous signaling compound. For example, if one was to identify 100 molecules that acted as inhibitors and all contained an aromatic ring, it is possible that the signaling molecule will also contain an aromatic ring. The information obtained from the unbiased library screening may provide clues as to the chemical methods of separation that could identify the signaling compound. Many universities now either own or have access to one or more of these small molecule libraries, and we are starting to see the identification of signaling inhibitors in many pathogens (Hentzer et al., 2003; Hung et al., 2005; Rasmussen et al., 2005; Muh et al., 2006a,b; Waters et al., 2008).
Whether one takes a biased or unbiased approach to screening for inhibitors there will be several key steps to validate the inhibitors identified by this process. Due to the large number of compounds that are going to be screened, it is likely that the assay will take place in either a 96- or 384-well plate format. The small reaction volumes make performing the assay in duplicate a requirement, as well as verifying that the assay is still functional in such small volumes. The addition of exogenous signaling molecule from either a synthesized source or from spent media will ensure that maximal signaling is possible by the reporter construct. The inclusion of reaction wells with no inhibitor, as the positive controls, and wells with media that do not contain any signaling molecule, as the negative controls, are essential in the ability to determine if inhibition is occurring. Each other reaction well will contain the strain with the reporter construct, added exogenous signaling molecule, and potential inhibitor. The determination of the concentration of the exogenous signaling molecule and potential inhibitor are key variables, and if the exogenous signal is weak the result may be a large number of inhibitors that are not potent. Conversely, if the concentration of the exogenous signal is too high relative to the inhibitors, then very few inhibitors will be identified. These ratios are key in determining the success of the screen and should be established prior to initiating the high-throughput screen.
Once inhibition has been observed, determining what a significant result is will prevent the further examination of compounds that may only have modest potency. In the EHEC example below, the standard variation of the assay was calculated, and then inhibitors that were altered >3 standard deviations from the mean of the control were considered significant. The compounds that were considered potential inhibitors were then collected from the various plates in the library and the assay was repeated to ensure that the results were reproducible. The compounds that were consistently inhibitory were selected and used to counterscreen. The counterscreen employs a non-CCS-activated promoter fusion construct, usually a promoter that is metabolic in nature or activated only under certain conditions. In the case of the specific example below, we utilized the β-lactamase promoter sequence. The counterscreen will identify compounds that generally inhibit the growth, general transcription inhibitors, and/or metabolism of the reporter strain and, as such, are not specific for the CCS system. This counterselection screen eliminates many potential inhibitors as potential therapeutics. While a general metabolic inhibitor may be desirable if we are looking for antibiotics, the purpose of this screen is to specifically inhibit the CCS-activated virulence mechanisms. This specific inhibition should result in less potential for the development of resistance, as the selective pressure of a CCS inhibitor will be much less than the selective pressure of the inhibition of growth (Fig. 17.3.2).
A well-designed and executed screen should provide multiple candidate compounds to be examined for therapeutic potential.
BASIC PROTOCOL
IDENTIFICATION OF E. COLI TEVS232 INHIBITORY COMPOUNDS VIA A HIGH-THROUGHPUT SCREENING
It is difficult to provide a protocol for every possible screen, so in this unit we will provide an example of the method used for the identification of a CCS inhibitor within the EHEC system. A flowchart of the steps taken in the screen is provided in Figure 17.3.3A and the detailed steps are described below. The reporter system developed for this assay is based on the introduction of the LEE1::lacZ fusion into the chromosome of a nonpathogenic E. coli isolate resulting in the strain E. coli TEVS232 (Sperandio et al., 1999). This strain will respond to the addition of a preconditioned medium from the pathogenic strain E. coli 8624. We optimized the growth of the reporter strain (variables in growth time, aeration, and medium were examined) and the standard growth protocol is as follows.
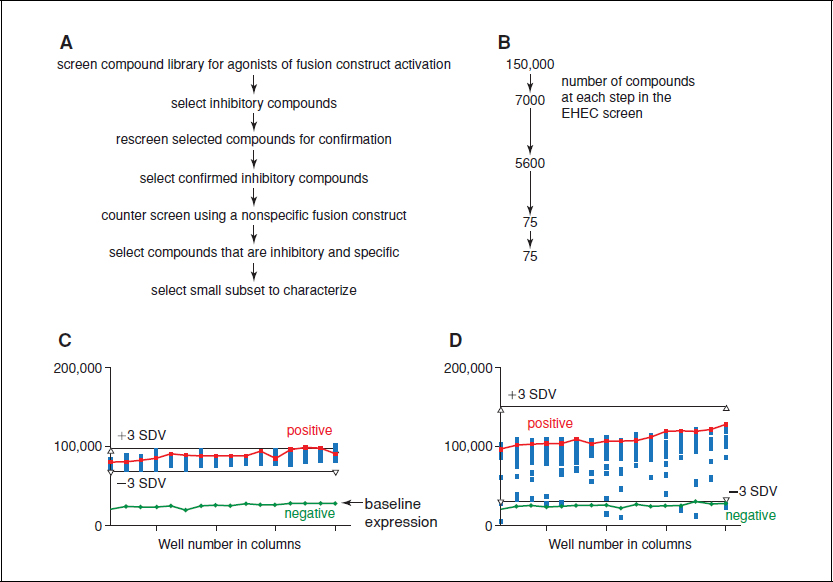
Figure 17.3.3 High-throughput screen for CCS-inhibitor identification. (A) A flowchart of the major steps in the high-throughput screen utilized in the identification of potential CCS inhibitors. (B) The number of potential EHEC CCS inhibitors identified at each step in A. (C) The results of a control plate in the EHEC CCS inhibitor screen. The green line is the negative control without preconditioned medium and thus represents the basal expression. The red line is the positive control in which preconditioned medium has been added. The upper black bar is three standard deviations above the mean of the control (+3 SDV) and the lower black bar is three standard deviations below (−3 SDV). (D) An experimental plate with compounds added to each of the wells in the 384-well plate. Each blue box represents a separate compound tested in the assay. The green, red, and black bars are the same as in Panel C. Compounds that are below the lower black bar (−3 SDV) and the basal level of expression were considered potential inhibitors and collected for further analysis.
The identification of the inhibitory compounds via a high-throughput screen provides a starting point for continued compound characterization. The compounds that are deemed acceptable inhibitors will have to be further tested to ensure that they do not inhibit the growth of the bacterium. The pressure to develop resistance against standard antibiotics is great, and it is our hope that the lack of growth pressure of the CCS inhibitors will allow them to be effective for a greater period of time or be used in combination with antibiotic therapies. In essence, the bacteria’s signaling is prevented from distinguishing the host from the environment and, as such, the bacteria will not activate virulence traits and will pass out of the host system with no detrimental effects.
Materials
E. coli TEVS232 (contains LEE1::lacZ chromosomal fusion)
Luria broth (Miller base; Invitrogen)
Dulbecco’s modified Eagle medium (DMEM; Invitrogen)
EHEC-preconditioned medium (see Support Protocol)
Dimethyl sulfoxide (DMSO)
Small molecule library (this study utilized the UT Southwestern Medical Center, Department of Biochemistry)
Lysozyme
Beta-Glo reagent (Promega)
37°C, 5% CO2 incubator
37°C shaking incubator
384-well plates
Biomek FX liquid handler
Additional reagents for growing E. coli in liquid medium (Elbing and Brent, 2002)
Stay updated, free articles. Join our Telegram channel

Full access? Get Clinical Tree