19.1 INTRODUCTION TO HEPADNAVIRUSES
The hepadnaviruses got their name because they cause hepatitis and they have DNA genomes. They are known as hepatitis B viruses (HBVs) and are classified in the family Hepadnaviridae. Some members infect mammals and some infect birds; examples include woodchuck hepatitis virus and heron HBV. The best known hepadnavirus is that which infects humans; it is commonly referred to as HBV, and is of major importance as an agent of disease and death. Duck HBV, on the other hand, is non-pathogenic in its natural host.
The hepadnaviruses are especially fascinating for two reasons. First they have very small genomes, which are used with great economy to encode the virus proteins and to control expression of the virus genes. Second, their DNA genomes are replicated via an RNA intermediate. In other words, their replication involves reverse transcription, so they are very different from DNA viruses that replicate their DNA directly to DNA. The discovery of the mode of replication of the hepadnavirus genome led to the creation of Baltimore Class VII.
DNA viruses that replicate via RNA have also been found in plants; an example is cauliflower mosaic virus. These viruses, together with the hepadnaviruses, have been termed pararetroviruses. This chapter will concentrate on HBV.
19.2 IMPORTANCE OF HBV
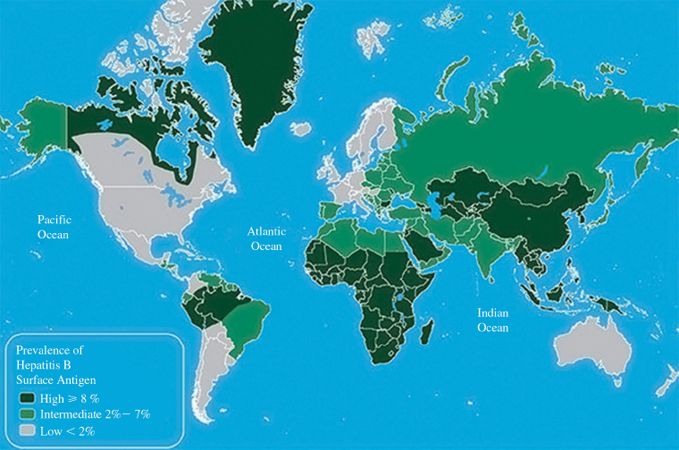
No one knows how many people are infected with HBV, but the figure is estimated to be around 350 million. The majority are in Asia and many are in Africa (Figure 19.1). There are high percentages of people infected in the far north of North America and Greenland, but because of the low populations of these regions the numbers infected are relatively small.
Figure 19.1 World distribution of HBV infection.
Source: Courtesy of the US Centers for Disease Control and Prevention.
Virus is present in the blood and semen of infected individuals and the modes of transmission generally parallel those for HIV transmission. There are over 50 million new HBV infections each year, the majority in babies who acquire the infection from their mothers. Many infections are thought to result from the reuse of syringes and needles for injections, mainly in the developing world.
Many HBV infections result in mild symptoms or are asymptomatic, especially in children. It is in children, however, that HBV infection is most likely to become persistent, with 90–95% of those infected as newborn infants becoming long-term carriers, compared with 1–10% of those becoming infected as adults.
Individuals who are persistently infected with HBV may remain healthy for much of the time, but some develop severe hepatitis, which may lead to cirrhosis and eventually to liver cancer (Chapter 23). These diseases resulting from HBV infection cause over half a million deaths each year.
19.3 HBV VIRION
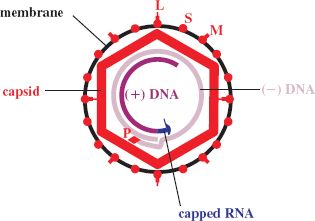
The virion is roughly spherical, with a diameter of about 42 nm. The main virion components are an envelope enclosing a capsid, inside which is the DNA and P (polymerase) protein (Figure 19.2).
19.3.1 DNA
The genome is made up of two strands of DNA, one of which is incomplete; hence the DNA is partly single-stranded and partly double-stranded. A short sequence is triple-stranded as a result of a complementary sequence at the 5′ ends, and this results in the DNA having a circular conformation (Figure 19.2). The genome is very small, with a length of about 3.2 kb(p).
At the 5′ end of each of the DNA strands there is a covalently linked molecule: a capped RNA on the short strand and a protein (P) on the long strand.
19.3.2 P (polymerase) protein
The virion contains at least one molecule of P (polymerase) protein. The N terminus of P constitutes a “terminal protein” domain; this is separated by a “spacer” from a domain with reverse transcriptase activity (Figure 19.3). At the C terminus there is a domain with ribonuclease H (RNase H) activity. P also has DNA-dependent DNA polymerase activity.
19.3.3 Capsid
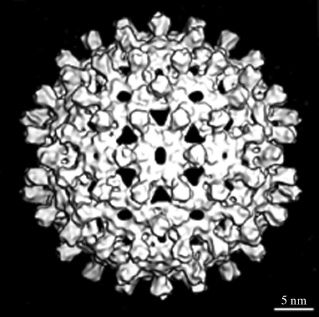
The capsid, which has icosahedral symmetry, has holes in it and short spikes protrude from its surface (Figure 19.4). It is constructed from dimers of the C (core) protein, which is largely α-helical, unlike the capsid proteins of many other viruses. The C terminus of the C protein is highly basic due to the presence of a large number of arginine residues; this region binds the virus genome.
Figure 19.4 HBV capsid. Derived from cryo-electron microscopy images of capsids assembled in E. coli cells expressing HBV C protein.
Source: Watts et al. (2002) The EMBO Journal, 21, 876. Reproduced by permission of Nature Publishing Group and the authors.
19.3.4 Envelope
The virion envelope contains three protein species designated as small (S), medium (M) and large (L). The M and L proteins are longer versions of the S protein (Figure 19.5), which is the most abundant of the three. Each protein has one or more glycosylation sites, though not all molecules are glycosylated. The surface regions of the envelope proteins constitute an antigen known as hepatitis B surface antigen (HBsAg).
Figure 19.5 Arrangement of HBV small (S), medium (M), and large (L) proteins in the virion envelope. The N termini of about 50% of L protein molecules are outside the membrane and about 50% are inside.
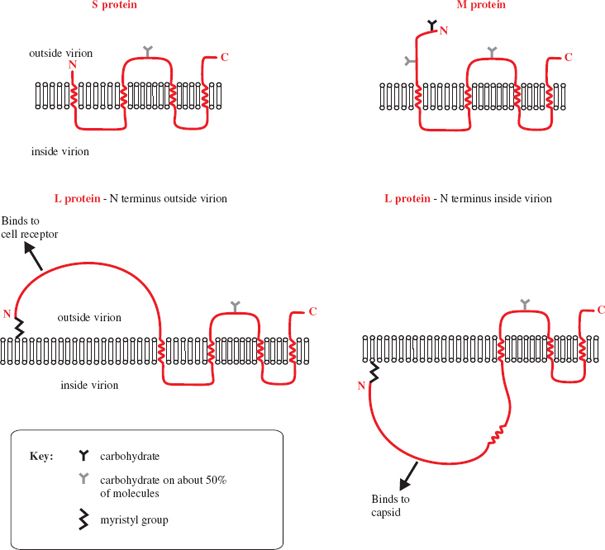
The binding site for cell receptors is near the N terminus of the L protein, but only about 50% of the L molecules have the N terminus on the outside of the virion; the N termini of the remaining L molecules are on the inside bound to the capsid.
19.4 NON-INFECTIOUS PARTICLES
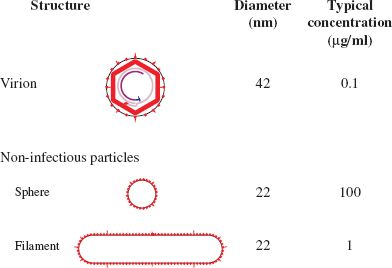
An unusual and intriguing feature of HBV infection is the presence in the blood of not only virions but also large quantities of non-infectious particles that have been released from infected liver cells (Figure 19.6). These particles are composed of lipid and virus envelope proteins, but they do not contain nucleocapsids. Some of the particles are spheres and some are filaments; both have diameters of 22 nm and the filaments have variable lengths up to 200 nm.
All the particles (virions and non-infectious particles) are much more abundant in the blood than in the liver, and the non-infectious particles, especially the spheres, vastly outnumber the virions. Presumably the virus has a good reason for investing in the production of such large amounts of non-infectious material, but that reason is not clear. The non-infectious particles may be a decoy for antibodies, thereby providing virions with some protection from the host’s immune system.
19.5 SOLUBLE VIRUS PROTEIN
In addition to the virions and non-infectious particles a soluble virus protein is found in the blood of some infected individuals. This protein is known as hepatitis B e antigen (HBeAg). It is similar to the C protein, but has 10 extra amino acid residues at the N terminus and lacks 34 amino acid residues at the C terminus. The function of HBeAg, like that of the non-infectious particles, may be to protect the virus from the immune response of the host.
19.6 HBV GENOME
At 3.2 kb, the HBV genome is very small, though there are viruses with smaller genomes (Section 3.2.1). There are four ORFs, from which seven proteins are translated (Figure 19.7), so a large amount of coding information is packed into the small genome. The virus cleverly achieves this by using every nucleotide in the genome for protein coding and by reading more than half of the genome in two reading frames. The P ORF, which occupies about 80% of the genome, overlaps the C and X ORFs and the S ORF is entirely within the P ORF. A further way in which the virus maximizes its coding capacity is by expressing the L protein in two different conformations that have different functions. In one conformation the L protein displays a site which can bind to a cell receptor, while in the other it binds the virion envelope to the capsid (Figure 19.5).
Figure 19.7 HBV genome organization. The incomplete (+) strand and the complete (–) strand of the DNA are shown surrounded by the four ORFs.
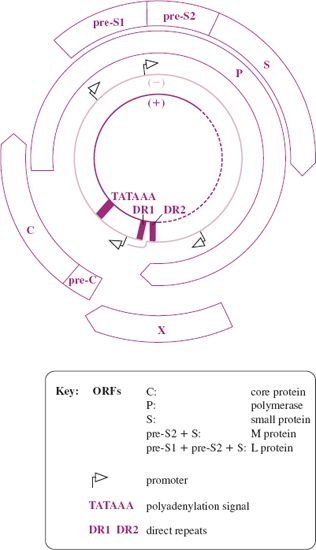
As the entire genome is involved in coding for protein, it follows that all the regulatory sequences, such as promoters, are within protein coding sequences. The genome contains direct repeats of 11 nucleotides known as DR1 and DR2.
Translation of the S region produces the S protein, translation of pre-S2—S produces the M protein and translation of the complete pre-S1—pre-S2—S ORF produces the L protein. Similarly, expression of the pre-C—C region gives rise to two proteins. Even though the virus maximizes the use of its small genome it encodes only seven proteins, so it is heavily dependent on host cell functions. Host proteins that have been demonstrated to play roles in the replication of HBV include enzymes, transcription factors, and chaperones.
19.7 HBV GENETIC GROUPS
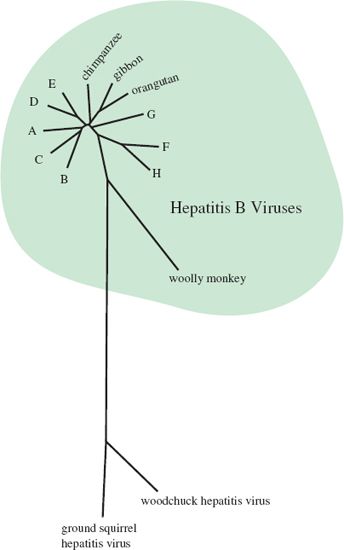
Sequencing of many HBV isolates has revealed eight genetic groups (genotypes A—H), and has demonstrated that these human viruses are related to similar viruses in other primate species (Figure 19.8).
Figure 19.8 Phylogenetic tree showing relationships between hepatitis B viruses from humans (genotypes A–H) and other primate species. Relationships with hepatitis viruses from two other mammalian species (woodchuck and ground squirrel) are also shown.
Stay updated, free articles. Join our Telegram channel

Full access? Get Clinical Tree