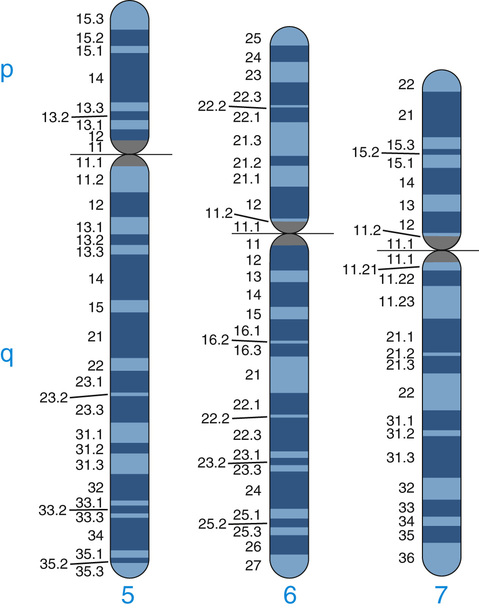
Human chromosomes are often classified into three types that can be easily distinguished at metaphase by the position of the centromere, the primary constriction visible at metaphase (see Fig. 5-2): metacentric chromosomes, with a more or less central centromere and arms of approximately equal length; submetacentric chromosomes, with an off-center centromere and arms of clearly different lengths; and acrocentric chromosomes, with the centromere near one end. A potential fourth type of chromosome, telocentric, with the centromere at one end and only a single arm, does not occur in the normal human karyotype, but it is occasionally observed in chromosome rearrangements. The human acrocentric chromosomes (chromosomes 13, 14, 15, 21, and 22) have small, distinctive masses of chromatin known as satellites attached to their short arms by narrow stalks (called secondary constrictions). The stalks of these five chromosome pairs contain hundreds of copies of genes for ribosomal RNA (the major component of ribosomes; see Chapter 3) as well as a variety of repetitive sequences.
In addition to changes in banding pattern, nonstaining gaps—called fragile sites—are occasionally observed at particular sites on several chromosomes that are prone to regional genomic instability. Over 80 common fragile sites are known, many of which are heritable variants. A small proportion of fragile sites are associated with specific clinical disorders; the fragile site most clearly shown to be clinically significant is seen near the end of the long arm of the X chromosome in males with a specific and common form of X-linked intellectual disability, fragile X syndrome (Case 17), as well as in some female carriers of the same genetic defect.
High-Resolution Chromosome Analysis
The standard G-banded karyotype at a 400- to 550-band stage of resolution, as seen in a typical metaphase preparation, allows detection of deletions and duplications of greater than approximately 5 to 10 Mb anywhere in the genome (see Fig. 5-1). However, the sensitivity of G-banding at this resolution may be lower in regions of the genome in which the banding patterns are less specific.
To increase the sensitivity of chromosome analysis, high-resolution banding (also called prometaphase banding) can be achieved by staining chromosomes that have been obtained at an early stage of mitosis (prophase or prometaphase), when they are still in a relatively uncondensed state (see Chapter 2). High-resolution banding is especially useful when a subtle structural abnormality of a chromosome is suspected. Staining of prometaphase chromosomes can reveal up to 850 bands or even more in a haploid set, although this method is frequently replaced now by microarray analysis (see later). A comparison of the banding patterns at three different stages of resolution is shown for one chromosome in Figure 5-4, demonstrating the increase in diagnostic precision that one obtains with these longer chromosomes. Development of high-resolution chromosome analysis in the early 1980s allowed the discovery of a number of new so-called microdeletion syndromes caused by smaller genomic deletions or duplications in the 2- to 3-Mb size range (see Fig. 5-1). However, the time-consuming and technically difficult nature of this method precludes its routine use for whole-genome analysis.
Fluorescence In Situ Hybridization
Targeted high-resolution chromosome banding was largely replaced in the early 1990s by fluorescence in situ hybridization (FISH), a method for detecting the presence or absence of a particular DNA sequence or for evaluating the number or organization of a chromosome or chromosomal region in situ (literally, “in place”) in the cell. This convergence of genomic and cytogenetic approaches—variously termed molecular cytogenetics, cytogenomics, or chromonomics—dramatically expanded both the scope and precision of chromosome analysis in routine clinical practice.
FISH technology takes advantage of the availability of ordered collections of recombinant DNA clones containing DNA from around the entire genome, generated originally as part of the Human Genome Project. Clones containing specific human DNA sequences can be used as probes to detect the corresponding region of the genome in chromosome preparations or in interphase nuclei for a variety of research and diagnostic purposes, as illustrated in Figure 5-5:
• DNA probes specific for individual chromosomes, chromosomal regions, or genes can be labeled with different fluorochromes and used to identify particular chromosomal rearrangements or to rapidly diagnose the existence of an abnormal chromosome number in clinical material.
• Repetitive DNA probes allow detection of satellite DNA or other repeated DNA elements localized to specific chromosomal regions. Satellite DNA probes, especially those belonging to the α-satellite family of centromere repeats (see Chapter 2), are widely used for determining the number of copies of a particular chromosome.
Stay updated, free articles. Join our Telegram channel

Full access? Get Clinical Tree


