and Sergey P. Laptenok2
(1)
Faculty of Sciences, Department of Physics and Astronomy, VU University Amsterdam, Amsterdam, The Netherlands
(2)
Laboratoire d’Optique et Biosciences, INSERM U696-CNRS UMR7645, Ecole Polytechnique, Palaiseau, France
Abstract
This chapter describes a procedure of global analysis of the steady-state spectra measured with different concentrations of the denaturant to quantitatively study protein denaturation. With the help of physicochemical models, relevant spectral parameters that characterize the folding intermediate and thermodynamic parameters that describe a three-state model N 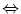 I
I 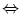 U can be estimated.
U can be estimated.
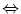 I
I 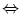 U can be estimated.
U can be estimated.Key words
Global analysisProtein denaturationSingular value decompositionSteady-state fluorescence1 Introduction
During denaturant-induced equilibrium (un)folding of a particular protein, a molten globule-like folding intermediate is formed [1]. Here we describe how the steady-state fluorescence spectrum monitored as a function of the denaturant concentration can be used to infer the properties of the folding intermediate with the help of global analysis [2].
2 Materials
Materials, Protein Expression, and Purification. All chemicals used were of the highest purity available. The concentration guanidine hydrochloride (GuHCl) was determined by measuring the refractive index of the sample used, as described previously [3]. A variant of apoflavodoxin from Azotobacter vinelandii, which contains two tryptophan residues (i.e., W74-W128-F167 (WWF)), was obtained and purified as described [3]. In all experiments protein concentration was 4 μM in 100-mM potassium pyrophosphate buffer, pH = 6.0. Temperature was set to 25 °C.
Steady-State Fluorescence Spectra. Steady-state fluorescence spectra were obtained with a Fluorolog 3.2.2 spectrofluorometer (Horiba, Jobin Yvon, Optilas, Alphen aan den Rijn, the Netherlands), as described previously [3]. The excitation wavelength was 300 nm, excitation and emission slit widths were 2 nm, and emission spectra were recorded between 305 and 400 nm with 1-nm steps. All spectra were corrected for wavelength-dependent instrumental response characteristics. Background fluorescence emission was measured under the same circumstances, except that now no protein is present in the samples, and was subsequently subtracted from the corresponding fluorescence spectra of samples with protein.
3 Methods
3.1 Determination of the Number of Components Contributing to the Steady-State Fluorescence Spectra
The steady-state spectra measured at 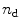 denaturant concentrations can be collated in a matrix
denaturant concentrations can be collated in a matrix 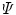 where a column of the
where a column of the 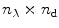 matrix
matrix 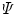 contains a spectrum measured at
contains a spectrum measured at 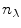 wavelengths at a particular denaturant concentration, whereas a row contains the emission measured at a particular wavelength at
wavelengths at a particular denaturant concentration, whereas a row contains the emission measured at a particular wavelength at 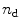 denaturant concentrations. The rank of this matrix
denaturant concentrations. The rank of this matrix 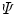 can be estimated with the help of the singular value decomposition (SVD) [4–11] (see Note 1).
can be estimated with the help of the singular value decomposition (SVD) [4–11] (see Note 1).
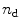 denaturant concentrations can be collated in a matrix
denaturant concentrations can be collated in a matrix 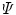 where a column of the
where a column of the 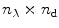 matrix
matrix 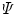 contains a spectrum measured at
contains a spectrum measured at 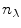 wavelengths at a particular denaturant concentration, whereas a row contains the emission measured at a particular wavelength at
wavelengths at a particular denaturant concentration, whereas a row contains the emission measured at a particular wavelength at 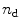 denaturant concentrations. The rank of this matrix
denaturant concentrations. The rank of this matrix  can be estimated with the help of the singular value decomposition (SVD) [4–11] (see Note 1).
can be estimated with the help of the singular value decomposition (SVD) [4–11] (see Note 1).Figure 1 depicts the results from the singular value decomposition (SVD) analysis of the data matrix  obtained from denaturant-induced unfolding of WWF apoflavodoxin. The scree plot of the singular values (Fig. 1b) shows a kink after i = 3 indicating the presence of at least three significant components [12]. The first three LSVs and RSVs (black, red, and blue in Fig. 1c, d) show clear structure. The fourth LSV and RSV (light green) are noise-like traces. In conclusion, SVD indicates that three species are present in the data matrix. There are several methods to resolve these species; most well known are soft modeling (e.g., Multivariate Curve Resolution (MCR) [13–15]) and hard modeling with the help of models that incorporate physicochemical knowledge and aim for the estimation of parameters that are physicochemically interpretable. The latter approach is termed global analysis [11, 16]. Advantages of hard over soft modeling have been demonstrated [13, 17]. A prerequisite for global analysis (see Note 2) is the availability of suitable physicochemical models.
obtained from denaturant-induced unfolding of WWF apoflavodoxin. The scree plot of the singular values (Fig. 1b) shows a kink after i = 3 indicating the presence of at least three significant components [12]. The first three LSVs and RSVs (black, red, and blue in Fig. 1c, d) show clear structure. The fourth LSV and RSV (light green) are noise-like traces. In conclusion, SVD indicates that three species are present in the data matrix. There are several methods to resolve these species; most well known are soft modeling (e.g., Multivariate Curve Resolution (MCR) [13–15]) and hard modeling with the help of models that incorporate physicochemical knowledge and aim for the estimation of parameters that are physicochemically interpretable. The latter approach is termed global analysis [11, 16]. Advantages of hard over soft modeling have been demonstrated [13, 17]. A prerequisite for global analysis (see Note 2) is the availability of suitable physicochemical models.
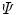 obtained from denaturant-induced unfolding of WWF apoflavodoxin. The scree plot of the singular values (Fig. 1b) shows a kink after i = 3 indicating the presence of at least three significant components [12]. The first three LSVs and RSVs (black, red, and blue in Fig. 1c, d) show clear structure. The fourth LSV and RSV (light green) are noise-like traces. In conclusion, SVD indicates that three species are present in the data matrix. There are several methods to resolve these species; most well known are soft modeling (e.g., Multivariate Curve Resolution (MCR) [13–15]) and hard modeling with the help of models that incorporate physicochemical knowledge and aim for the estimation of parameters that are physicochemically interpretable. The latter approach is termed global analysis [11, 16]. Advantages of hard over soft modeling have been demonstrated [13, 17]. A prerequisite for global analysis (see Note 2) is the availability of suitable physicochemical models.
obtained from denaturant-induced unfolding of WWF apoflavodoxin. The scree plot of the singular values (Fig. 1b) shows a kink after i = 3 indicating the presence of at least three significant components [12]. The first three LSVs and RSVs (black, red, and blue in Fig. 1c, d) show clear structure. The fourth LSV and RSV (light green) are noise-like traces. In conclusion, SVD indicates that three species are present in the data matrix. There are several methods to resolve these species; most well known are soft modeling (e.g., Multivariate Curve Resolution (MCR) [13–15]) and hard modeling with the help of models that incorporate physicochemical knowledge and aim for the estimation of parameters that are physicochemically interpretable. The latter approach is termed global analysis [11, 16]. Advantages of hard over soft modeling have been demonstrated [13, 17]. A prerequisite for global analysis (see Note 2) is the availability of suitable physicochemical models.
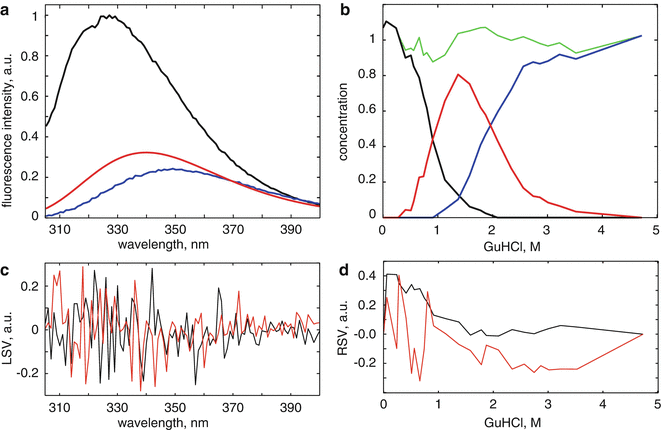
Fig. 1
Results from the singular value decomposition (SVD) analysis of the data matrix collated from steady-state fluorescence data obtained from denaturant-induced unfolding of WWF apoflavodoxin. (a) Denaturation trajectory of steady-state fluorescence spectra obtained at increasing concentrations of denaturant. (b) Scree plot of the singular values shows a kink after i = 3 indicating the presence of at least three significant components. (c) The first four left singular vectors (LSVs) (colored black, red, blue, and light green, respectively). (d) The accompanying first four right singular vectors (RSVs)
3.2 Global Analysis of the Steady-State Fluorescence Spectra with the Help of a Spectral Model
At each GuHCl denaturant concentration, the observed emission spectrum 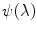 is described as a linear combination of spectra arising from native protein (n), folding intermediate (i), and unfolded protein (u):
is described as a linear combination of spectra arising from native protein (n), folding intermediate (i), and unfolded protein (u):
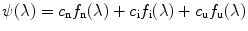
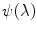 is described as a linear combination of spectra arising from native protein (n), folding intermediate (i), and unfolded protein (u):
is described as a linear combination of spectra arising from native protein (n), folding intermediate (i), and unfolded protein (u):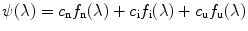
(1)
Steady-state fluorescence spectra obtained of protein at 0 and 4.72 M GuHCl are used as reference spectra that characterize the native and unfolded protein, respectively. The steady-state fluorescence spectrum of the folding intermediate was modeled as a skewed Gaussian in the energy domain (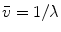 ) and is described by three parameters: peak location
) and is described by three parameters: peak location 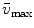 , width
, width 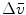 , and skewness b [16, 11]:
, and skewness b [16, 11]:
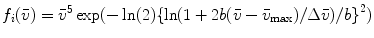
where the parameter 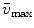 is the Franck-Condon wave number of maximum emission. The full width at half maximum (FWHM) is given by
is the Franck-Condon wave number of maximum emission. The full width at half maximum (FWHM) is given by 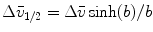 (see Note 3).
(see Note 3).
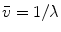 ) and is described by three parameters: peak location
) and is described by three parameters: peak location 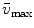 , width
, width 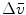 , and skewness b [16, 11]:
, and skewness b [16, 11]: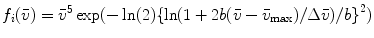
(2)
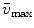 is the Franck-Condon wave number of maximum emission. The full width at half maximum (FWHM) is given by
is the Franck-Condon wave number of maximum emission. The full width at half maximum (FWHM) is given by 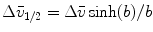 (see Note 3).
(see Note 3).Now all spectra are globally analyzed as a linear combination of spectra arising from native protein, folding intermediate, and unfolded protein. The three parameters that describe the shape of the spectrum of the folding intermediate and the concentrations of each folding species are the unknown parameters that need to be estimated from a global fit of all data. The 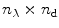 matrix
matrix 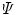 can be written as a matrix product.
can be written as a matrix product.
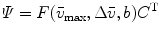
where the F matrix contains three columns ![$$ \left[ {\begin{array}{*{20}{c}} {{f_{\mathrm{ n}}}} & {{f_{\mathrm{ i}}}({{\bar{v}}_{\max }},\Delta \bar{v},b)} {{f_{\mathrm{ u}}}} \\\end{array}} \right] $$](/wp-content/uploads/2017/03/A299540_1_En_3_Chapter_IEq000319.gif) and
and 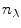 rows and the C matrix contains three columns
rows and the C matrix contains three columns ![$$ \left[ {\begin{array}{*{20}{c}} {{c_{\mathrm{ n}}}} & {{c_{\mathrm{ i}}}} {{c_{\mathrm{ u}}}} \\\end{array}} \right] $$](/wp-content/uploads/2017/03/A299540_1_En_3_Chapter_IEq000321.gif) and
and 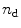 rows. The rank of the C, F, or
rows. The rank of the C, F, or 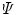 matrix is three, consistent with the SVD. The concentration parameters were constrained to be nonnegative (see Note 4). The estimated spectrum of the folding intermediate is depicted in red in Fig. 2a. Note that it is blue shifted relative to that of the unfolded protein (blue). The estimated spectral parameters are
matrix is three, consistent with the SVD. The concentration parameters were constrained to be nonnegative (see Note 4). The estimated spectrum of the folding intermediate is depicted in red in Fig. 2a. Note that it is blue shifted relative to that of the unfolded protein (blue). The estimated spectral parameters are 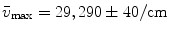 ,
, 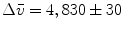 , and
, and 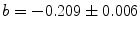 . The root mean square error of the fit was 0.41 % of the maximum of the data. The matrix of residuals resulting from the global analysis can best be diagnosed with the help of its SVD. Shortcomings of the model used show up as trends in the most important left or right singular vectors. No such trends are present in the first (black) and second (red) LSV or RSV (Fig. 2c, d). Therefore, the fit can be accepted.
. The root mean square error of the fit was 0.41 % of the maximum of the data. The matrix of residuals resulting from the global analysis can best be diagnosed with the help of its SVD. Shortcomings of the model used show up as trends in the most important left or right singular vectors. No such trends are present in the first (black) and second (red) LSV or RSV (Fig. 2c, d). Therefore, the fit can be accepted.
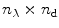 matrix
matrix 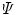 can be written as a matrix product.
can be written as a matrix product.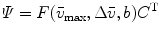
(3)
![$$ \left[ {\begin{array}{*{20}{c}} {{f_{\mathrm{ n}}}} & {{f_{\mathrm{ i}}}({{\bar{v}}_{\max }},\Delta \bar{v},b)} {{f_{\mathrm{ u}}}} \\\end{array}} \right] $$](/wp-content/uploads/2017/03/A299540_1_En_3_Chapter_IEq000319.gif) and
and  rows and the C matrix contains three columns
rows and the C matrix contains three columns ![$$ \left[ {\begin{array}{*{20}{c}} {{c_{\mathrm{ n}}}} & {{c_{\mathrm{ i}}}} {{c_{\mathrm{ u}}}} \\\end{array}} \right] $$](/wp-content/uploads/2017/03/A299540_1_En_3_Chapter_IEq000321.gif) and
and  rows. The rank of the C, F, or
rows. The rank of the C, F, or 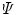 matrix is three, consistent with the SVD. The concentration parameters were constrained to be nonnegative (see Note 4). The estimated spectrum of the folding intermediate is depicted in red in Fig. 2a. Note that it is blue shifted relative to that of the unfolded protein (blue). The estimated spectral parameters are
matrix is three, consistent with the SVD. The concentration parameters were constrained to be nonnegative (see Note 4). The estimated spectrum of the folding intermediate is depicted in red in Fig. 2a. Note that it is blue shifted relative to that of the unfolded protein (blue). The estimated spectral parameters are 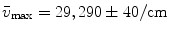 ,
, 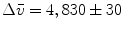 , and
, and 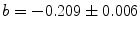 . The root mean square error of the fit was 0.41 % of the maximum of the data. The matrix of residuals resulting from the global analysis can best be diagnosed with the help of its SVD. Shortcomings of the model used show up as trends in the most important left or right singular vectors. No such trends are present in the first (black) and second (red) LSV or RSV (Fig. 2c, d). Therefore, the fit can be accepted.
. The root mean square error of the fit was 0.41 % of the maximum of the data. The matrix of residuals resulting from the global analysis can best be diagnosed with the help of its SVD. Shortcomings of the model used show up as trends in the most important left or right singular vectors. No such trends are present in the first (black) and second (red) LSV or RSV (Fig. 2c, d). Therefore, the fit can be accepted.
Fig. 2




Global analysis of steady-state fluorescence data matrix with the help of a spectral model. (a) Steady-state fluorescence spectra of WWF apoflavodoxin in 0 M GuHCl (native protein, black) and in 4.72 M GuHCl (unfolded protein, blue), respectively. The steady-state fluorescence spectrum of the folding intermediate (red) is modeled as a skewed Gaussian and is estimated from the global analysis of all unfolding data. (b) Estimated concentrations of the different folding species (colored black, red, and blue) as a function of denaturant concentration. The sum of the three concentrations is shown as a thin light green line. (c, d) Results from the singular value decomposition of the residual matrix. (c) The first (black) and second (red) LSV. (d) The first (black) and second (red) RSV
Stay updated, free articles. Join our Telegram channel

Full access? Get Clinical Tree


