Ideally, the developed signal model should be validated against template signals.1 This validation could be performed based on, for example, statistical parameters such as sum of square error (SSE), correlation between modelled signal and template signal, execution time, and so on. The best-fit modelling method can then be chosen. Alternatively, a visual inspection could be performed to evaluate the fit of the model against the template signal. However, if a very accurate model is required, then both approaches should be performed.
Once the developed signal model has been validated against template signals, and thus can imitate the real phenomena, it means that the original values of the signal model parameters (P1O, P2O, P3O, …PnO) are set. These values will only be modified in the simulator by the end-user.
Now, it is required to build a corresponding simulator where the predefined signal model parameters (P1O, P2O, P3O, … PnO) are also possibly controlled (i.e. overwritten) by the end-user. Moreover, other parameters (internal to the adaptation process) could also be introduced in the simulator by the end-user; these can also be controlled by the end-user. These other parameters are used inside the adaptation process to tune the signal model parameters in order to reflect the actual phenomena that take place in the biological system/object of interest.
The core mechanisms of the simulator include adaptation. Either the adaptation of the signal model is done according to the end user’s need/requirement or his/her will to simulate the signals. The generator generates the simulated signals as per end-user’s prescribed parameters (P1G, P2G, P3G, … PnG), so that the end-user is able to control the signal model parameters and generate the simulated signals as desired.
The simulated signals could be used for further analysis or evaluation of the performance of algorithms, e.g. separation algorithms.
The above block diagram of a generic system (Fig. 3.1) and a generic framework (Fig. 3.2) for modelling bioelectrical information that can guide the model and development process has been devised. The framework serves to measure the parameters of interest for the biological system/object and the process to clean the measured data in order to achieve the ideal (template) of signals (Mughal et al. 2016).
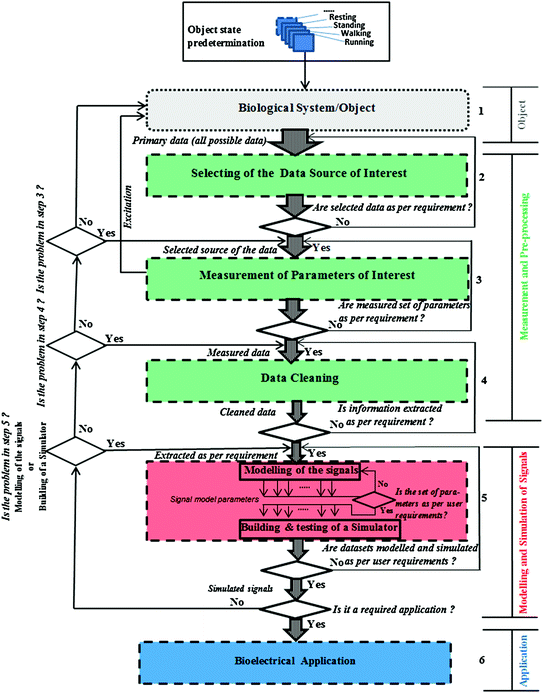

Fig. 3.2
Flow diagram of the proposed novel generic framework for modelling and simulation the bioelectrical information (Mughal et al. 2016)
The flow diagram of the processes according to the proposed framework is depicted in Fig. 3.2. This flow chart guides the advanced user step by step with the help of the predefined blocks (Mughal et al. 2016).
Each diagram has specific criteria which must be kept in mind and guidelines that must be followed.
The details of this generic framework are discussed in what follows.
3.1 Novel Generic Framework
Figure 3.2, a flow diagram of data acquisition, processing, and modelling and simulation of the bioelectrical information is given.
This flow diagram is the pathway to application in order to model the signals. In the next chapter (see Sect. 4.4.1 in Chap. 4), this flow diagram is implemented for the specific case of EBI based on the IRG and ICG signals.
In the first step (Step 1 in Fig. 3.2), a biological system or an object is selected; in the second step (Step 2), the data source of interest is selected; in the third step (Step 3), the parameters in which the advanced user (e.g. technical measurement personnel) is interested are measured (i.e. those which are directly measurable). After measuring the parameters of interest, data cleaning techniques are applied in the fourth step (Step 4). Then, after cleaning the data, the signals are modelled and a corresponding simulator is built in the fifth step (Step 5). In the sixth and final step (Step 6), the application of interest is developed as per the end user’s needs (Mughal et al. 2016).
3.2 Detailed Explanation of Each Step of the Novel Generic Framework
In this section each step is discussed in detail, from the first step (Step 1, selection of biological system/object) to the last step (Step 6, bioelectrical application of the novel generic framework).
3.2.1 Description of the Biological System/Object (Step 1)
This diagram (Fig. 3.3) contains three systems, namely the cardiovascular system, the respiratory system, and the muscular system; each of these is illustrated by a sub-diagram. Each system contains parameters (which are the most interesting in this work), and these parameters are connected with each other within the same system. Some of these parameters are also connected with parameters which are contained in one or both of the two other systems.
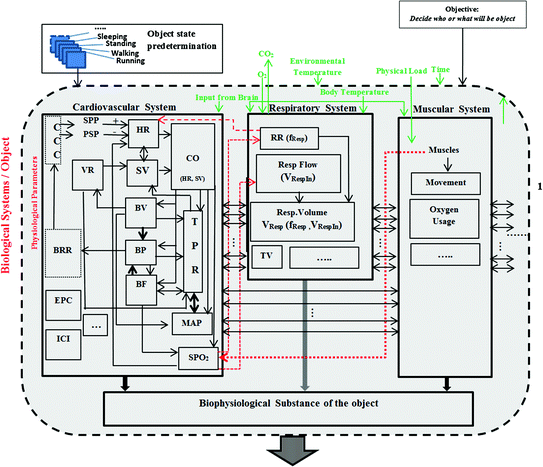

Fig. 3.3
Block diagram of the relationship of parameters of three main systems (cardiovascular, respiratory, and muscular). It shows the connection/relationship/dependency between the parameters inside each system or between the systems (Mughal et al. 2016)
Connection/relationship/dependency between the parameters inside the system or outside the system are shown with different arrows as explained below.
Thin arrows (solid line): show the dependency to some extent on the other parameter(s) inside the same system.
Thick arrows (solid line): show the direct relation, strong dependency on the other parameter(s) inside the same system.
Thin arrows (dotted line): show the dependency to some extents on the other parameter(s) within another system.
Thick arrows (dotted line): show the direct relation, strong dependency on the parameter(s) within another system.
Thick arrows (both site direction): show the direct proportional to each other with the same system (Mughal et al. 2016).
The following description starts from the BRR and follows the natural flow of the three systems.
3.2.1.1 Parameters of the Cardiovascular System
BRR (Baroreceptor Reflexes): Blood pressure (BP) is controlled on a minute-to-minute basis by BRR. Changes in BP affect the frequency of action potentials sent to the Cardiovascular Control Centre (CCC) from the BRR. BRR is discussed previously in Chap. 2 in Sect. 2.1 and in Timischl 1998.
CCC (Cardiovascular Control Centre): Heart Rate (HR) is controlled by both sympathetic (SPP) and parasympathetic (PSP). SPP (Sympathetic) increase the HR. PSP (Parasympathetic) decrease the HR.
HR (Heart Rate): HR corresponds to the frequency of heartbeat, i.e. the number of heartbeats per minute or reciprocal of the duration of heart cycle Yc = 1/Tc [beats/min].
VR (Venous Return): VR is the amount of blood that returns to the heart (VR dependent on BV (Blood Volume)) expressed in time units [l/min].
SV (Stroke Volume): SV is the volume of blood which is pumped out by the heart with a single beat (SV is dependent on VR and Total Peripheral Resistance (TPR)). The HR and SV are proportional to each other [l/min].
CO (Cardiac Output): CO is the volume of blood which is pumped out by the heart per minute. It is a function of HR and SV (CO is dependent on HR and SV) [beats/min].
BP (Blood Pressure): BP usually refers to the arterial pressure of the systemic circulation. It is partly dependent on CO and the vessels, and directly (strongly) depends on BV and BF (Blood Flow)). In Sect. 2.1 of Chap. 2 and Timischl 1998 it is discussed in detail [mmHg].
BV (Blood Volume): BV is the volume of blood (both red blood cells and plasma) in the circulatory system of any individual. It is dependent on CO.
TPR (Total Peripheral Resistance): the blood vessels provide resistance to the flow of blood. The resistance and pressure are directly (strongly) proportional to each other. If the resistance increases, then the pressure increases (TPR is dependent on CO, BV, BP, BF and MAP) [Ohms].
BF (Blood Flow): The flow of the blood through the vessels of the circulatory system is a function of the BP and TPR (BF is dependent on BP, TPR, and CO). In Sect. 2.1 of Chap. 2 and Timischl 1998 it is discussed in detail [l/min].
MAP (Mean Arterial Pressure): MAP represents the average driving force for the blood flow through the arterial system (MAP is dependent on CO, BV, and directly proportional with TPR) [mmHg].
SPO 2 (Saturation Pressure of Oxygen): The muscles highly depend on SPO2 because if the muscle starts to work, they require more oxygen (SPO2 is dependent on muscles, CO, and BF) [Percentage].
ICI (Intra–Cardiography Impedance): Measures the cardiac output internally [Ohms].
EPG (Epicardial Potential): Internal ECG [V].
3.2.1.2 Parameters of the Respiratory System
RR (Respiration Rate): RR is the number of cycles per minute. It is not directly dependent on HR, but under certain conditions, it is dependent on HR and SPO2 [cycles/min].
RF (Respiration Flow): Inspiration or expiration volume of airflow in a minute. RF is also dependent on SPO2 [l/min].
RV (Respiration Volume): RV is the volume of air that is inhaled and exhaled per minute. It is a function of RR and RF (RV is dependent on the RR and the RF) [l/min].
TV (Tidal Volume): TV is the volume of gas inhaled or exhaled during one respiratory cycle. It is discussed in detail in Sect. 2.4 of Chap. 2 and Krivošei 2009.
Stay updated, free articles. Join our Telegram channel

Full access? Get Clinical Tree


