Fig. 17.1
Working principle of a biosensor. The analyte is specifically recognized by the bioelement. A biochemical reaction occurs upon the event of recognition leading to the generation of a signal, which can be amplified. Exposure of a biosensor to a non-specific analyte leads to no recognition signal
Recognition Elements
In a biosensor, different types of bioelement can be used. In this section some of them will be described without the ambition of completeness. For more details on bioelements, the reader is referred to excellent reviews by Chambers et al. [5] and Bhalerao and Nistala [6]. Figure 17.2 shows some of the different types of bioelement. The main requirement for a good bioelement is that it is highly specific for the analyte with a high affinity. The affinity of the bioelement to the analyte is the limiting factor for the sensitivity of a biosensor [7].
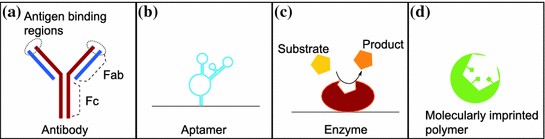

Fig. 17.2
Four different bioelements for a biosensor. a Antibodies are widely used in biosensors and other immunoassay due to the availability of antibodies against of wide range of targets. b Aptamers are oligonucleotides that specifically bind and recognize various targets. c Enzyme based biosensors have been extensively investigated. The high specificity of the enzyme for its target makes it very attractive for biosensor applications. d Molecular imprinted polymers (MIP) is a bioelement that is synthesized upon polymerisation of monomers around the target that can be separated leaving an imprint that can catch the target with a very high specificity
Enzymes
Widely used types of bioelement are enzymes. Enzymes are highly selective for their substrate (i.e. the analyte) and they catalyse the binding event. After binding of the substrate on the active sites, a chemical reaction occurs producing a measureable product, and releasing the enzyme for further reactions [8]. Often the enzyme-based biosensors are coupled with an electrochemical transducer. This is the case for example in case of enzyme-based glucose biosensors. The glucose oxidase (GOx) oxidizes glucose to gluconolactone, thereby using oxygen and producing H2O2 [9]. The consumption of oxygen can be measured by an oxygen electrode or the production of H2O2 can be measured amperometrically. The main disadvantage of enzymes is the cost and the time consuming purification that is required to have a concentration of enzymes high enough in the system.
Antibodies
The second—and probably most widely used—bioelements for analyte recognition in biosensor are antibodies. Antibodies are proteins produced by the immune system of many organisms. Their function is to recognize and neutralize pathogenic agents in a highly specific way. Due to their high specificity, each antibody molecule will recognize only one defined target. Antibodies are composed of two heavy chains and two light chains forming a “Y” having two antigen recognition sites (at the Fab regions) and a constant region (Fc region) as shown on Fig. 17.2.
Antibodies are produced by immunizing an animal (mouse, rabbit, horse, etc.) with the target. After some days, the serum of the animal will contain antibodies in high concentration against the injected target. The serum is collected and purified by affinity chromatography in order to obtain an antibody solution that is qualified as polyclonal, since it will contain several types of antibodies against one target (each type of antibody binding to a different binding site of the target.). Polyclonal antibodies solutions are relatively cheap to produce and find application amongst others in enzyme-linked immunosorbent assays (ELISA). However they might lack specificity for certain biosensors [6]. The specificity is improved by using monoclonal antibodies, which are mixtures of antibodies containing only one set of antibody against one particular binding site of the target. They are produced using the hybridoma technology where immortalized cancer cells are fused with antibody-producing B-cells in order to obtain a cell line producing one single set of antibody. With the progress of the recombinant technology, antibodies have also been developed having only the antigen-binding region (Fab fragment) and also antibodies carrying Fab and Fc fragments from different organisms. For biosensors, the Fc region is as well of great importance as it can be used for enhancement of the detection signal. Antibodies against the Fc region of a e.g. mouse antibody can be produced in a different organism (e.g. rabbit). Different markers (fluorescent molecules, biotin, gold particles, etc.) can then be conjugated to the Fc part of this second antibody that permits the detection of the binding event. This type of sandwich assay is widely used in ELISA and lateral flow assays like the pregnancy test.
Aptamers
Aptamers represent an attractive alternative to antibodies as bioelements. They are short ssDNA or RNA oligonucleotide sequences (20–40 bases) that fold into a 3D-structure that has a high selectivity and affinity for a defined target [10]. The 3D-structure is stabilized after binding to the target. Aptamers are smaller in size compared to antibodies and they can be synthesized chemically so that no batch-to-batch variation is introduced into the biosensor. Thank to this chemical synthesis, aptamers can easily be modified with alternative groups and conjugated to any kind of markers either at the 3′ or at the 5′ end of the sequence [11]. Aptamers are thermally stable as they can resist several cycles of denaturation/renaturation [11].
Aptamers can be developed against any type of target (metal ion, biomolecules, cells, etc.) [10]. The selection is usually run in a cycle called Systematic Evolution of Ligands by Exponential Enrichment (SELEX) [12]. A random pool of different aptamers is incubated with the target. The aptamers bound to the target are separated from the non-bound aptamers. After release of the target, the binding aptamers are driven through a new selection cycle. This process is repeated until the affinity of the aptamers to the target is high enough (dissociation constant in the nano- to picomolecular range). Recently, advancements in the SELEX technology have greatly reduced the time and costs required for the selection of an aptamer. This in vitro selection gives the possibility of selecting aptamers against non-immunogenic and toxic substances for which no antibodies can be developed [13].
Aptamer-based biosensors, also called aptasensor have gain a wide interest in the last years due to the advantages of aptamers compared to antibodies. Similar to antibodies, a variety of immobilization methods is available to bind aptamers to the sensor element. Aptasensors can be coupled to an electrochemical, optical or mass-sensitive transducer [13]. One of the successful examples for aptasensor was the detection of thrombin which was widely investigated [14]. Xiao et al. [15] have made an interesting development: a redox compound (methylene blue) was inserted into the thrombin aptamer. When the target bound to the aptamer, the induced conformation change inhibited the electron transfer from the methylene blue to the electrode. This change could be detected amperometrically.
Molecular Imprints
Going even more towards synthetic recognition bioelements, we would like to draw attention to molecularly imprinted polymers (MIP) that were first investigated by Mosbach and Wulff in the 1970s [16]. MIP will be described more in detailed in Sect. “Molecularly Imprinted Polymers (MIP): An Alternative to Bioelement”. Briefly, during the polymerization the monomers of the polymer are self-assembling around the target. After removing the target, an imprint of the target is remaining, which can be used as recognition element for the biosensor. MIP offer a greater chemical stability than biomolecules [17] and can be developed for most of the targets.
Other Recognition Elements
Further bioelements that are worth mentioning here are whole cells (eukaryotic cells or bacterial cells) such as the fibroblast cells which were used by Feng et al. [18] to develop a biosensor for the electrochemical measurement of the ATP concentration. Another type of biosensor widely used is the DNA hybridization sensor. In this type of sensor, a DNA probe is immobilized on the transducer of the sensor. When the complementary strand hybridizes to the probe, a signal is generated that is then detected. As an example, the sensor developed by Nascimento et al. [19] detected bovine papillomavirus DNA electrochemically on a sensor functionalized with a specific probe of 27 bases.
Sensitivity and Selectivity of Biosensors
Sensitivity and selectivity are used to evaluate the biosensors. In the physical meaning, the sensitivity indicates how much the signal changes with a change of analyte concentration, while the selectivity shows how good the sensor can detect the analyte among the complex matrix of the sample (for example in blood, urine, saliva etc.). The limit of detection (LOD) is also often a measure of the quality of the sensor as it determines the lowest concentration of analyte that can still be detected by the sensor, i.e. gives a significant change in the sensor response. When the analyte is detected without any further labelling by a second probe, the literature refers to the sensors as label-free sensors [7]. Biosensors often are label-free sensors.
To increase the sensitivity and specificity of the biosensors in most cases the bioelement is immobilized on the active part of the biosensor. This process is called “functionalization”. Without aiming to give a full list of methods the reader can find some of the most used processes for immobilization in the followings.
Functionalization
Several immobilization methods are available in order to functionalize the biosensor [6].
Absorption of the bioelement onto the surface of the biosensor is often used. This is a simple method since it only requires the incubation of the bioelement on the surface, but the amount of active bioelement on the surface is not well controlled [20]. Various types of non-covalent interactions can take place during the adsorption of the bioelement such as attractive Van-der-Waals forces, electrostatic interactions, and hydrophobic interactions [6, 21].
Covalent binding is often the most desirable methods of binding the bioelement as it is more controlled and reliable [6]. Covalent binding can be achieved using various linkers like 1-ethyl-3-(3-dimethylaminopropyl) carbodiimide hydrochloride (EDC) [22] or glutaraldehyde on materials containing OH-groups on the surface. Thiol group modified bioelements can bind to a gold surface. Silanes can be used to bind the bioelements on glass or on silicon.
The bioelement can also be incorporated into a porous material. In this case the analyte must be able to diffuse freely into the material in order to reach the bioelement. This offers a high surface-to-volume ratio but controlling the concentration of the bioelement is difficult. Enzymes are often entrapped in such a porous material.
Important is however that the immobilization does not affect the structure of the sites for analyte recognition as well as the function of the bioelement.
Transducer
Once the target is captured by the recognition element, the binding must generate a signal that can be detected by a transducer. The biosensing event can be only qualitative (analyte is there or not) or also quantitative (how much of the analyte is present in the tested sample). Several transduction methods are available, among others: optical, electrochemical or mechanical [23].
Optical Transducers
Optical methods require that a light or colour signal is produced by the recognition event. This could be for example light produced by an enzyme reaction. Fluorescence signalling is also often used in biosensors. Optical transduction can also be done in the form of a colorimetric assay, where the binding event results in a colour change of the recognition layer [24]. In lateral flow assays, nanoparticles are conjugated to the antibodies. During the recognition, the colour of the particles is changing, in most cases forming a coloured line on the sensor.
Electrochemical Transducers
Electrochemical transducers are also often integrated in biosensors. An extensive development has enabled the systems to become sensitive and highly specific for the substrates. The signal can easily be processed and analysed. The working principle of the electrochemical transducer is that the binding event can induce a change in the electrical properties of the system. Different types of devices are available [25]. If electroactive species are oxidized or reduced during the biochemical reaction, a current can be measured. This is an amperometric detection, which can be divided in two modes: amperometry when the potential is kept constant and voltammetry when the potential is varied [26]. Since the biomolecules are often not electroactive, a mediator can be added to the reaction chain, which will—in most cases—increase the sensitivity [26]. In potentiometry, the potential between working and reference electrode is measured when no current flows. By that, the ion activity of the sample can be measured, which is related to the concentration of analyte [27]. In the course of the biochemical reaction, the charges in the system are changed (for example: due to the production of electrons in an enzymatic reaction) [26, 28]. This change is then monitored. Conductometric sensors measure the change in the electrode resistance due to the binding event on the surface. Often AC voltage with small amplitude (10–20 mV) is used for measuring the impedance of an electrode system in impedimetric sensors. These sensors detect the changes of impedance upon changes on the electrode surface and/or in the solution [29]. Since the binding event is detected at the surface of the electrode, it has to occur in close proximity of this electrode; that is within the Debye length. The small size of aptamers meets this requirement and makes them a very attractive bioelement for electrochemical-based biosensors.
Other Transducers
Other transduction methods include mass-sensitive methods like the cantilever-based sensors [30]. Cantilevers are usually made of small silicon plate (length: 100 µm) with a low force constant. They are fixed at one end and can vibrate at the other end. Silicon can easily be functionalized with bioelements such as antibodies or aptamers. Upon binding of the analyte to the bioelement at the surface of the cantilever, the resonance frequency of the cantilever is changing, which can be detected with help of a laser beam shining the cantilever surface [31].
To summarize, biosensors are very versatile devices. There is a broad range of possible configurations regarding bioelements for analyte recognition, as well as for the signal transducer. In the next section we describe how the polymers are used for the development of biosensors.
Conductive Polymers in Biosensors
Polymers as Device Materials
Biosensors made of plastic materials emerged because of their favourable properties: cost effectiveness, suitability for mass replication based on injection moulding, excellent stability providing inexpensive, high-throughput, and large-scale devices with environmentally friendly disposability.
Several biocompatible polymers are known and used for device fabrication. The most commonly used materials are poly(methyl methacrylate) (PMMA), Cyclic olefin copolymer (COC) and polydimethylsiloxane (PDMS).
Recently several methods were developed for fast prototyping and mass production of micro- and nanostructures in polymers. Injection moulding is one of the techniques, which is commonly used with great success. Improvement in patterning, structuring [32] or bonding [33] of those materials is essential for the creation of low cost and portable biosensors. However, in this work, we do not focus on these fabrication methods, rather we describe some specific polymers which are commonly used in biosensor designs.
Polymers are mostly known for their insulating properties and their roles as dielectrics and resists [34]. However, a class of polymers can conduct electricity, and therefore they are called “conductive polymers” (CP). The use of CPs as electrode materials is becoming more and more widespread for biological applications, especially in the field of biosensors [35], as they are cheaper and easier to produce than conventional metal electrodes.
Conductive Polymers as Electrode Materials
Polyaniline was the first CP polymer, which was described in the mid-19th century by Henry Letheby [36]. Since then numerous intrinsically CP have been developed, among others: polyacetylene, polythiophene, polypyrrole. CPs, also referred to as synthetic metals, have found applications in many fields. They are integrated for example in solar cells, rechargeable batteries and biomedical devices [37]. CPs are also very attractive for biosensors. In biosensors, CP can be used as excellent non-metallic electrodes. Numerous biosensors have been developed over the past 20 years with electrodes made of CP. The fabrication is fairly easy and flexible. This allows the biosensors to be single-use system; avoiding any risk of contamination and adaptation of the biosensors to new targets can be rapidly made. They are mostly biocompatible, can easily be synthesized and can be modified for immobilization of bioelements [38]. These conductive polymers are referred to as intrinsic conductive polymers in comparison to extrinsic conductive polymers that are a polymer matrix in which some metal particles have been entrapped [39].
Doping
The conducting process in polymers is rather complex. For the conducting process, the charge carriers have to be able to move in the material. This is achieved because the conjugated backbone of the polymer provides delocalized pi-orbitals that overlap continuously to allow free movement of the electrons [35, 40]. Besides delocalized electrons, the conductive polymers can have other charge carriers, which are often produced by doping with electron acceptors or donors [37]. The concept of polarons, bipolarons and solitons has been introduced to describe the charge defects introduced by the doping [41, 42]. Oxidation of the polymer creates holes (p-doping) while reduction adds electrons (n-doping) [34]. Different methods of doping can be applied [37]. For example, through chemical doping with iodine, the conductivity of polyacetylene could be increased over seven orders of magnitudes [37, 43, 44]. This discovery was rewarded the Nobel Prize in chemistry in 2000. Electrochemical doping can also be performed by the electropolymerization of the monomers. With this method, the ions come from the electrolyte solution [34]. The advantage of this method is that a polymer film is directly created on the electrode [45]. Photodoping introduces charge defects by irradiation of light [37].
In the following, three examples of conductive polymers will be discussed because these are the most commonly used in biosensors. Table 17.1 shows the chemical structures of polypyrrole, polyaniline and PEDOT, the three conductive polymers reviewed in this section.
Table 17.1
Chemical structure of polypyrrole, PEDOT, polyaniline
Polypyrrole | 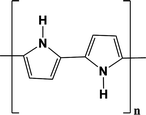 |
PEDOT | 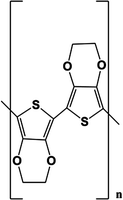 |
Polyaniline | 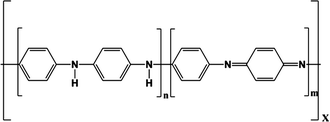 |
Polyaniline
Polyaniline is a CP that has been known and used for a very long time. The synthesis of polyaniline, often abbreviated as PANI, is based on the oxidation of the monomer aniline. PANI has been reported to have a high conductivity (up to 103 S/m) [46, 47]. Thanks to its properties, it has found a wide range of sensing applications, inclusive in biosensors. The reader is referred to the excellent review of Dhand et al. [48]. Horseradish peroxidase (HRP) is an enzyme extensively incorporated in biosensors. It catalyzes the reduction of hydrogen peroxide (H2O2), a reactive oxygen species, which is produced by a wide range of enzymatic reactions [49]. Detection of H2O2 is highly relevant in food and pharmaceutical industry. Chairam et al. [50] developed an enzyme based amperometric biosensor for the detection of H2O2. HRP was immobilized in the layer of conducting polymer poly(aniline-co-o-aminobenzoic acid) via electrostatic interactions. After mixing of the enzyme in the polymer, the mixture was simply cast onto a glassy carbon electrode. A layer of the biopolymer chitosan was then coated onto the HRP-p (Ani-co-o-Aba). This layer was used as a binder for the enzyme. This sensor showed a linear range from 10 to 1,000 µM with a detection limit of 1.8 µM. This novel and fast fabrication method showed excellent promising results for highly sensitive biosensors.
The biopolymer chitosan has also been used in combination with PANI films by Srivastava et al. [51]. They developed a modified PANI-gold electrode with immobilization of cholesterol oxidase on a chitosan matrix. First, nanocomposites of PANI and gold were prepared. After complete polymerisation, the nanocomposites were incorporated in chitosan. This mixture was then spin coated onto an indium tin oxide (ITO) glass. The success of the immobilization was assessed by cyclic voltammetry measurements that showed a lower current peak caused by the electrical hindrance of the immobilized enzyme. The biosensor was optimized for the detection of cholesterol. Cholesterol is oxidized by cholesterol oxidase which produces H2O2, that is oxidized producing electrons that are transferred to the electrodes via the redox mediator Fe(CN)6−3. The signal is a peak in current that increases with the cholesterol concentration. The biosensor was stable over the time and retained a 90 % cholesterol oxidase activity over two weeks.
Composites of gold and PANI on a glassy carbon electrode were also used by Xiang et al. [52] for the development of a glucose biosensor. They used a bienzymatic approach where glucose was oxidized thereby producing H2O2, which was reduced by cytochrome oxidase c, absorbed on the surface of the nanocomposites. This sensor enabled a direct electron transfer to the electrode material. The sensor has a detection limit of 0.01 mM.
A similar electrochemical based sensor was developed using nanocomposites of PANI, graphene and horse radish peroxidase on ITO glas [53]. This biosensor was able to measure very low concentration of anti-malaria drug in various pharmaceutical preparations and in different body fluids (urine, plasma and serum).
Similarly, nanocomposites of graphene and PANI were used by Ruecha et al. [54] to develop a paper based biosensor for the enzyme based detection of cholesterol and the electrochemical detection of the by-product H2O2 of the enzymatic reaction. Interestingly they incorporated polyvinylpyrrolidone in the modified electrode in order to improve the stabilization of the graphene concentration. Those biosensors based on composites of different materials often show a combination of the parameters of the materials and therefore offer a higher thermal stability, better conductivity and improved resistance to salts.
Tamer et al. [55] worked on the optimization of the fabrication of the composites PANI/gold nanorod for glucose detection. In this biosensor, the enzyme GOx was covalently bound to the electrode via the linker glutaraldehyde. This matrix offered a large surface to functionalize.
PANI has also been shown to be usable in combination with whole bacterial cells as bioelement. Anu Prathap et al. [56] developed a biosensor where recombinant E. coli bacteria were immobilized by adsorption into a PANI film. The bacteria had been previously transformed with the gene linA2, coding for an enzyme involved in the first step of the degradation of pesticide lindane. Using recombinant E. coli bacteria assured a higher robustness and better performances to the sensor compared to the wild type which has a slow degradation rate of lindane. The sensor was proved to be functional during two weeks if stored at 4 °C. It could detect lindane in the part per trillion concentrations. It was highly sensitive and selective since it could detect the three isomers of lindane but showed no response for two other common pesticides. The advantage of using a whole cell as bioelement is to overcome the lack of synthetic bioelement for certain targets and provide the sensor with a high specificity. The detection was based on the change of conductivity induced in the PANI film after production of HCl by the enzymatic reaction. Amperometric measurements were used to register the increase in current upon production of HCl.
PANI was also used in combination with carbon nanotubes to fabricate a sensor with cytochrome oxidase as bioelement [57]. This system could be used for different kinds of sensing events based on the enzymatic reduction of H2O2. The sensitivity of a biosensor based on a PANI film on a gold electrode was improved by using a composite of PANI with polypyrrole nanoparticles [58]. The sensor was used to detect glutamate in food products with an enzymatic reaction of the covalently bound glutamate oxidase.
Nanowires based biosensors offer a high sensitivity. Lee et al. [59] developed a biosensor based on a single PANI nanowire that was functionalized via the EDC and N-Hydroxysuccinimide (NHS) (EDC/NHS) chemistry with antibodies against immunoglobulin G (IgG) and Myo, two cardiac biomarkers. The nanowire was fabricated using the electrochemical growth method between two gold electrodes. The binding of either IgG or Myo was recorded by the increase of conductance of the wire. The PANI biosensor was highly selective and sensitive to its target with detection limits of 3 ng/mL for IgG and 1.4 ng/mL for Myo.
Polypyrrole
Polypyrrole (PPy) is a conducting polymer that has attracted high interest in the biosensor field. PPy forms a yellow film that that becomes darker upon polymerization. PPy is indeed biocompatible, enabling the CP for the application in the medical biosensor field and shows a high conductivity [42]. It is worth mentioning that PPy has recently found application in the drug delivery field [60]. The synthesis of PPy has been extensively studied and can be done chemically or by electropolymerization. The way that the synthesis is performed has a significant effect on the properties of the PPy film [42, 61]. High conductive polypyrrole can reach a conductivity of up to 200 S/cm. Doped PPy films are relatively stable in air. The degradation of polypyrrole films starts with the loss of dopants, followed by the degradation of the polymer backbone [62]. Polypyrrole films have been described as being planar chains along the direction of the film layer.
Xu et al. [63] recently presented a biosensor for thrombin sensing based on polypyrrole electrodes prepared by electropolymerization on platinum electrodes. They used pyrrole-nitrilotriacetic acid (pyrrole-NTA) monomers that were polymerized and complexed with Cu2+. As bioelement, they immobilized a histidine labelled aptamer on the polypyrrole film through the histidine-NTA-Cu2+ interaction. The surface of this biosensor was shown to be non-adherent to proteins. The change of impedance was measured upon binding of the target to the aptamer. Thrombin was detected in the picomolar range (4.4 × 10−12 mol/L). Detection of the E. coli strain K12 was achieved by a DNA-based biosensor [64]. Short DNA sequences (25 base pairs) called hybridization probes were designed complementary to the sequence of the gene D-glucoronidase from E. coli. The DNA sequences were incorporated in the bulk of pyrrole that was electropolymerized onto a platinum electrode. The hybridization was measured by cyclic voltammetry with a clear difference between the amplitudes of the oxidation peaks resulting from the hybridization of a complementary sequence or from a non-complimentary sequence. The response of the biosensor could be obtained within 15–20 min which was much faster than any detection system involving culturing of the bacteria. Glucose biosensors have also been extensively investigated. They mainly use an enzyme-based system with GOx as bioelement and electrochemical detection. Olea et al. [65, 66] developed a new way to immobilize the enzyme GOx onto the biosensor surface. GOx was encapsulated in a multilamellar lipid vesicle before being incorporated into the polymer film. The comparison between free enzyme and encapsulated enzyme showed that the sensitivity was improved fivefold and that the stability of the biosensor was increased as well [67].
Stay updated, free articles. Join our Telegram channel

Full access? Get Clinical Tree


