CHAPTER 21 Molecular studies in myeloproliferative and myelodysplastic/myeloproliferative neoplasms
Introduction
Almost 60 years after William Dameshek published his description of myeloproliferative disorders,1 clinicians, pathologists, and basic scientists have noticed tremendous progress in the field of these diseases. Due to discovery of molecular markers, it is now possible to discriminate between myeloproliferative and reactive states. Targeted therapies for certain subtypes of myeloproliferative diseases have become available.
Discovery of the so-called Philadelphia (Ph+) chromosome in 1960 by Nowell and Hungerford,2 the subsequent dissection of its molecular structure and of pathways involved in t(9;22) translocation have led to the introduction of imatinib mesylate as the first molecularly targeted therapy in a human malignancy.3 Until 2005, knowledge of molecular aberrations and genetic defects in the Philadelphia-chromosome negative (Ph−) chronic myeloproliferative disorders (CMPD) was rather sparse. In a minority of cases, chromosomal changes such as trisomies 8 and 9, aberrations of chromosome 1, del(20q) or del(13q) in primary myelofibrosis (PMF) or loss of heterozygosity (LOH) at 9p in polycythemia vera (PV) have been demonstrated.4,5
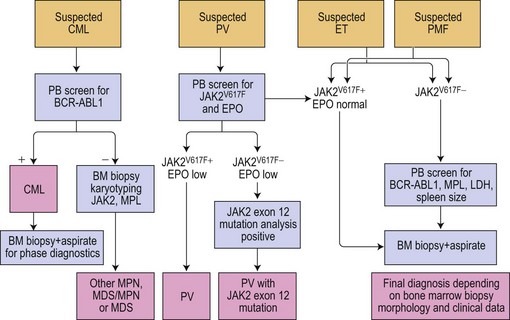
The discovery of the gain-of-function mutation V617F in the tyrosine kinase (TK) Janus kinase 2 (JAK2V617F)6 and other molecular defects in a considerable number of patients with so-called classical Ph− CMPD resulted in a revision of the WHO classification.7 To underline the neoplastic nature of these diseases, the WHO classification published in 2008 replaced the term Ph− CMPD with ‘myeloproliferative neoplasm (MPN)’. This new classification has also introduced new standards for diagnostic algorithms including molecular diagnostics (Fig. 21.1). In this chapter, the current knowledge on molecular changes in MPN and myelodysplastic/myeloproliferative neoplasms (MDS/MPN) is summarized. Molecular aspects of MPN with eosinophilia and of systemic mastocytosis are described in Chapters 25 and 26, respectively.
The Philadelphia chromosome and BCR-ABL in chronic myelogenous leukemia
Chronic myelogenous leukemia (CML) is a clonal stem cell disorder characterized by increased autonomous proliferation of myeloid lineages in the bone marrow (BM). The underlying molecular defect, the balanced reciprocal translocation t(9;22)(q34;q11), was the first chromosomal anomaly consistently present in a human cancer.8 Translocation t(9;22)(q34;q11.2) results in the Philadelphia chromosome found in 90–95 % of patients.9,10 This translocation causes a fusion of the BCR gene from chromosome 22 with the ABL1 gene from chromosome 9.11,12 In 5–10% of patients, a cryptic translocation of 9q34 and 22q11.2 or a variant translocation involving other chromosomes not detectable by conventional karyotype analysis occur. In these patients, the BCR-ABL1 fusion can be demonstrated by molecular methods such as fluorescent in situ hybridization (FISH), reverse transcriptase (RT)-PCR, and Southern blot.13
The BCR-ABL1 fusion results in a deregulated function of the ABL1 gene.14 BCR-ABL1 mRNA encodes for p210 or (rarely) p190 or p230 oncoproteins.15–19 The binding of ATP to the pockets of p210, p190 or p230 allows phosphorylation of selected tyrosine residues on its substrates. The oncogenic properties of the p210, p190, and p230 proteins rely primarily on the constitutively activated TK.
ABL1 plays a central role in the regulation of the cell cycle, cellular response to genotoxic events, and integrin-signaling pathways.20 The fusion of BCR with the SH3 domain of ABL1 interrupts the physiologic suppression of the kinase activity, produces the neoplastic phenotype via deregulation of the impact of ABL1 on apoptosis, cell cycle, and cellular adhesion pathways.21,22 Activation of the RAS pathway results in an increase of mitotic activity and up-regulation of bcl-2, while activation of ‘signal transducers and activators of transcription’ (STAT) 1 and STAT5 proteins results in the inhibition of apoptosis.23–25 Apoptosis inhibition counteracts the protective effect of ABL1 when DNA damage occurs. This results in the accumulation of neoplastic cells and additional genetic changes. Furthermore, BCR-ABL1 negatively influences DNA repair and drives centrosomal hypertrophy, thus increasing the risk of clonal evolution.26,27 BCR-ABL1 induces abnormalities of cytoskeletal cellular function, phosphorylation of adhesion proteins, the production of an abnormal variant of the β1-integrin, and the phosphorylation of Cdc2-related kinase (Crk)-l resulting in reduced adhesion of the CML cells to BM stroma.28–30 Interrupted by BCR-ABL1 interaction between hematopoietic progenitor cells and the BM stroma appears to be important for the control of cellular proliferation.
Nowadays, it is generally accepted that CML originates from a single BCR-ABL1 transformed pluripotent hematopoietic stem cell.31 This immature precursor and its immature daughter cells appear to be rather resistant to therapy utilizing BCR-ABL1 TK inhibitors. Therefore, this type of anti-neoplastic treatment does not seem to cure the disease.32
BCR-ABL1 positive cells or small positive clones have been reported in asymptomatic persons without any signs of CML.33 Moreover, there are differences in the risk of CML between patients with different HLA-B phenotype.34 Thus, immunologic factors may also play a role in the evolution of CML, probably restricting the occurrence of CML to subjects with an immunologic failure of resistance against the BCR-ABL1 positive cell population.
Imatinib mesylate and second generation formulas such as nilotinib and dasatinib represent TK inhibitors that have been designed to work against the constitutively activated chimeric BCR-ABL1 protein, which is responsible for the aberrant phenotype of affected cells in CML patients.35,36
Karyotyping and conventional cytogenetics accompany the inevitable histopathological evaluation of BM biopsy in the initial diagnostic algorithm in order to demonstrate the presence of the Ph+ chromosome. Detection of the BCR-ABL1 fusion products by reverse transcriptase (RT)-PCR analysis in patients with CML (and also Ph+ ALL) has become the golden standard in the diagnostic setting. Standardized real-time quantitative (RQ)-PCR has become the methodology of choice for monitoring molecular response to treatment with TK inhibitors and/or interferon.37 The achievement of the major molecular response (MMR) is the key issue in evaluating therapy effects. MMR can only be determined by quantitative RT-PCR methodology and was originally defined as the reduction of the BCR-ABL1 fusion transcripts by three logs below a standardized baseline level. The latter was initially introduced as a part of the International Randomized Study of Interferon versus STI571 (IRIS) trial.38 Since most patients treated with imatinib or other TK inhibitors achieve a complete cytogenetic response (CCyR), the monitoring of minimal residual disease (MRD) by quantification of the BCR-ABL1 transcripts has become increasingly important. It has been shown that CCyR and MMR after 12 months of treatment indicates an improved progression-free survival at 24 months in 100% of the patients treated with imatinib as compared to 95% in the group showing no MMR and 85% in patients who were not in CCyR at 12 months.38 All patients who achieved CCyR and MMR at 18 months had a 5-year progression-free survival. Moreover, studies in patients followed for several years showed that those achieving MMR early in the first year of treatment had significantly longer CCyR39.
Despite the improved prognosis of CML patients since start of the imatinib era, a response failure is observed in 25–30% of patients.40 The individual risk of developing resistance to imatinib is hard to predict and underlying mechanisms are largely unknown. The majority of patients develop de novo mutations in the ABL kinase domain during therapy. Some may even have pre-existing mutations hindering a good response to initial treatment.
A study of lymphoid blast crisis of CML has recently shown that lymphoid blasts but not CML cells express a B-cell specific mutator named AID (antibody diversification enzyme activation-induced deaminase), which promotes genetic instability and somatic hypermutation of tumor suppressor and DNA repair genes.41 AID may also be responsible for the acquisition of imatinib resistance in CML cells, which justifies testing for the potential targeting of AID and further stresses the need for BCR-ABL1 transcript level monitoring during TK inhibitor treatment.
Due to the clinical importance of MRD, efforts to standardize the methodologies for quantification of BCR-ABL1 transcripts in CML patients are now in progress. Most studies refer to the IRIS trial and the associated International Scale (IS) for BCR-ABL1 transcript quantification,38 but significant inter-laboratory differences concerning definition of patient molecular status still exist. The increase of BCR-ABL1 transcripts to a level >0.1% on the IS scale can be interpreted as a potential response failure and should lead to a close-meshed monitoring of BCR-ABL1 transcript levels.42 Successful efforts to standardize methodologies have already been reported.43 The goal of a recently initiated study is to introduce a conversion factor (CF) allowing the harmonization and comparison of results obtained during molecular monitoring of CML patients before and during therapy.44 In brief, every participating laboratory generates its own CF, which will allow comparison of BCL-ABL1 transcript levels for upcoming multicenter studies.
Interestingly, there is no concordance between BCR-ABL1 transcript levels in either sorted or unsorted BM aspirate cells and the levels determined in peripheral blood cells, both mononuclear cells and granulocytes. Since drawing blood is less invasive as compared to BM aspiration, it should be considered the standard choice to determine the BCR-ABL1 transcript levels. Apart from BCR-ABL1 levels, other prognostic parameters such as the presence of BM fibrosis45 or increasing numbers of BM blasts are of importance during clinical follow-up (see Chapter 24 for details).
The Janus kinase 2 in MPN
JAK2V617F
The molecular nature of the gain-of-function mutation in JAK2 is a hotspot in exon 14 where the wild-type (WT) guanine (G) is changed to a mutant thymine (T) with consecutive replacement of valine by phenylalanine at position 617 (V617F) in the JH2 pseudokinase.6,46–48 Even though functional studies are not yet available, it is well accepted that the bulky amino acid phenylalanine changes the conformation of the protein thereby blocking the interaction of the auto-inhibitory JH2 pseudokinase domain with the catalytic domain JH1. This hampers control of catalytic activity and leads to a constitutively activated JAK2 kinase and subsequently, the activation of downstream targets in affected cells.49
The effects of mutated JAK2V617F show a considerable diversity in various cellular lineages and MPN subtypes affected by this mutation. The JAK2V617F clone is autonomous and highly proliferative in vitro, even in the absence of growth factors. All MPN subtypes harboring the JAK2V617F show high numbers of endogenous erythroid colonies (EEC) in BM cultures. In addition, the affected lineages are additionally hypersensitive to growth factors such as erythropoietin (EPO), granulocyte-colony stimulating factor (G-CSF) or interleukins. In mouse models, the mutation is powerful enough to mediate a PV-like phenotype and leads to development of myelofibrosis.47,50,51 Similar findings were demonstrated in MPN patients harboring the JAK2V617F mutation, but some of the features observed in the animal model might be due to toxic effects of the JAK2V617F overdose. It is of note that strain-specific differences were found in two different mice models showing the JAK2V617F mutation. Whereas both the Balb/C and the C57Bl/6 mice showed an increase in hemoglobin and hematocrit, the number of leukocytes and the degree of myelofibrosis were strikingly higher in the Balb/C mice.50 No thrombocytosis was seen in these models, even though an increase of megakaryocytes in the BM was noted. Direct transfer of knowledge concerning JAK2V617F mutation derived from mice models to human MPN appears to be difficult and different phenotypes observed in humans may also develop as a consequence of individual host modifiers.
Among MPN, PV showed the highest JAK2V617F frequency (~95%) followed by PMF and ET (~50% JAK2V617F mutated cases).52–55 It is of note that patients with JAK2V617F mutated ET show PV-like features, e.g. elevated hemoglobin and hematocrit levels. Also, it has been suggested that PV patients with a homozygous mutation status (i.e. 50% or more alleles showing the mutant T allele) had a higher risk for development of myelofibrosis.56–58 However, the mutation per se is probably not responsible for myelofibrosis development, since no correlation between the JAK2 status and the clinical course or degree of fibrosis could be demonstrated in PMF.59,60 A retrospective analysis of 490 MPN patients along with evaluation of follow-up biopsies revealed a correlation between JAK2V617F mutant allele burden and MPN subtypes. In this study, a considerable number of ET cases were characterized by lower allele burden.61
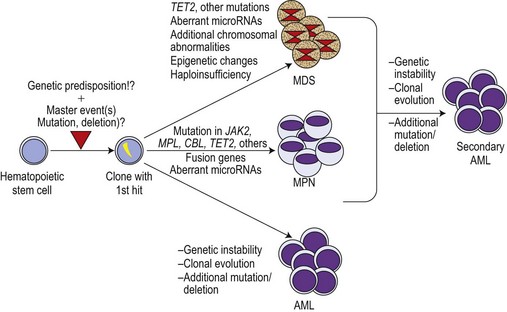
Although the discovery of the JAK2V617F mutation was a breakthrough in the field, it became increasingly clear that this molecular defect is not the initiating event in MPN pathogenesis. This is illustrated by X-chromosome inactivation patterns (XCIP) in female ET patients with the JAK2V617F mutation.62–64 XCIP studies found that the proportion of JAK2V617F positive cells was lower than the total number of clonal cells in a given patient. A higher proportion of cells with the deletion of chromosome 20q in comparison to the percentage of cells with the JAK2V617F mutation was found in patients carrying both anomalies.64 These studies clearly indicate that the neoplastic hematopoietic stem cell (HSC) must have acquired at least one molecular aberration preceding the JAK2V617F mutation. This master event could be an as yet undiscovered small chromosomal deletion or an insertion spanning only small-size region(s) of a gene, thereby impossible to detect by conventional cytogenetic techniques (Fig. 21.2). This event must lead to a selective advantage of the affected cell, allowing autonomous expansion accompanied by ongoing chromosomal instability and additional molecular defects such as mutations inducing entity-typical phenotypes. In MPN, the mimicry of phenotypes between different entities is not uncommon. This might be a result of two or more aberrations existing in parallel. It is therefore now widely accepted that JAK2V617F is a secondary mutation leading towards PV or a PV-like clinical appearance, particularly in ET.
JAK2 (K539L and other aberrations in exon 12)
In the diagnostic setting, some patients show high hemoglobin levels, high hematocrit and low serum EPO levels but do not harbor the JAK2V617F mutation. These patients are usually affected by a rather rare molecular defect in exon 12 of the JAK2 gene. Exon 12 aberrations vary between point mutations leading to the K539L mutation and insertions/deletions leading to various amino acid substitutions.65 Two interesting epidemiologic features characterize JAK2 exon 12 aberrations: 1) more women than men were affected when compared to idiopathic erythrocytosis showing the JAK2WT; 2) patients presented at younger age when compared to those with JAK2V617F mutation.66 However, due to the low frequency of JAK2 exon 12 aberrations and the complexity of testing, this type of molecular analysis should be restricted to patients who are JAK2V617F negative but show clinical parameters strongly suggesting clonal erythropoiesis.
The JAK2V617F mutation in de novo acute leukemia and in MPN transformed to acute leukemia
MPN transformation into acute myeloid leukemia (AML) is the major life-threatening event in the course of the disease. The frequency of transformation varies among the MPN subtypes. The general risk for evolving to AML is much higher for patients with PV or PMF than for those having ET. The impact of a history of prior treatment, i.e. with hydroxyurea, is unclear.67,68 The mechanism of the transforming switch is totally unknown. After discovery of the JAK2V617F mutation, this molecular defect has been suggested to be a powerful co-factor in the process of transformation. A series of studies investigated the occurrence of JAK2V617F in secondary AML developing in patients with a history of MPN and in de novo AML or acute lymphoblastic leukemia (ALL). In one study the JAK2V617F mutation was not detected in de novo AML M0 – M6-subtype (according to the French-American-British (FAB) classification) but some positive cases of AML M7 were found (2/11, 18%).69 Another investigation described one case of AML M6 (1/53) positive for JAK2V617F but no positive cases in AML M5 (0/85) or AML M7 (0/14) FAB categories.70 In one study a frequency of almost 3.0% was reported (3/113 de novo AML) with two AML cases showing both the JAK2V617F and t(8;21)(q22;q22). One case assigned to the ‘AML without maturation’ category had a JAK2 (V607N) mutation.71 In B- and T-ALL, the JAK2 mutation seems to be extremely rare, except for children with Down syndrome.72,73
To investigate the role of JAK2V617F in the transformation of MPN, the mutant allele burden (‘gene dosage’) has been retrospectively studied in individual patients. It was found that patients with a prior history of MPN and detectable JAK2V617F at diagnosis showed no increase of mutant allele burden during transformation to acute leukemia.74 The same study demonstrated that both the development of myelofibrosis and leukemic transformation in PV and PMF was found in patients with the JAK2WT status.
It has been noted that in JAK2V617F positive patients, the populations of blasts in transformed MPN are frequently negative for the JAK2 mutation.63 These findings are in line with the suggestion of the pre-JAK2V617F phase, in which clonality has already been achieved by an as yet undefined mechanism (see Fig. 21.2). Thus, JAK2V617F-negative AML developing during course of MPN might arise from the primary clone and could be the key to discovery of the primary master hit. The other scenario could be that, indeed, two separate clones develop independently in an individual patient. Studies of the molecular signatures of JAK2V617F-negative blasts from the AML transformation of MPN could open up relevant insights into the pathogenesis of early MPN development.
Stay updated, free articles. Join our Telegram channel

Full access? Get Clinical Tree