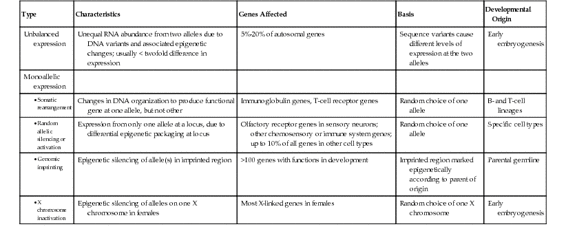
Monoallelic Gene Expression
Some genes, however, show a much more complete form of allelic imbalance, resulting in monoallelic gene expression (see Fig. 3-11). Several different mechanisms have been shown to account for allelic imbalance of this type for particular subsets of genes in the genome: DNA rearrangement, random monoallelic expression, parent-of-origin imprinting, and, for genes on the X chromosome in females, X chromosome inactivation. Their distinguishing characteristics are summarized in Table 3-2.
Somatic Rearrangement
A highly specialized form of monoallelic gene expression is observed in the genes encoding immunoglobulins and T-cell receptors, expressed in B cells and T cells, respectively, as part of the immune response. Antibodies are encoded in the germline by a relatively small number of genes that, during B-cell development, undergo a unique process of somatic rearrangement that involves the cutting and pasting of DNA sequences in lymphocyte precursor cells (but not in any other cell lineages) to rearrange genes in somatic cells to generate enormous antibody diversity. The highly orchestrated DNA rearrangements occur across many hundreds of kilobases but involve only one of the two alleles, which is chosen randomly in any given B cell (see Table 3-2). Thus expression of mature mRNAs for the immunoglobulin heavy or light chain subunits is exclusively monoallelic.
This mechanism of somatic rearrangement and random monoallelic gene expression is also observed at the T-cell receptor genes in the T-cell lineage. However, such behavior is unique to these gene families and cell lineages; the rest of the genome remains highly stable throughout development and differentiation.
Random Monoallelic Expression
In contrast to this highly specialized form of DNA rearrangement, monoallelic expression typically results from differential epigenetic regulation of the two alleles. One well-studied example of random monoallelic expression involves the OR gene family described earlier (see Fig. 3-2). In this case, only a single allele of one OR gene is expressed in each olfactory sensory neuron; the many hundred other copies of the OR family remain repressed in that cell. Other genes with chemosensory or immune system functions also show random monoallelic expression, suggesting that this mechanism may be a general one for increasing the diversity of responses for cells that interact with the outside world. However, this mechanism is apparently not restricted to the immune and sensory systems, because a substantial subset of all human genes (5% to 10% in different cell types) has been shown to undergo random allelic silencing; these genes are broadly distributed on all autosomes, have a wide range of functions, and vary in terms of the cell types and tissues in which monoallelic expression is observed.
Parent-of-Origin Imprinting
For the examples just described, the choice of which allele is expressed is not dependent on parental origin; either the maternal or paternal copy can be expressed in different cells and their clonal descendants. This distinguishes random forms of monoallelic expression from genomic imprinting, in which the choice of the allele to be expressed is nonrandom and is determined solely by parental origin. Imprinting is a normal process involving the introduction of epigenetic marks (see Fig. 3-8) in the germline of one parent, but not the other, at specific locations in the genome. These lead to monoallelic expression of a gene or, in some cases, of multiple genes within the imprinted region.
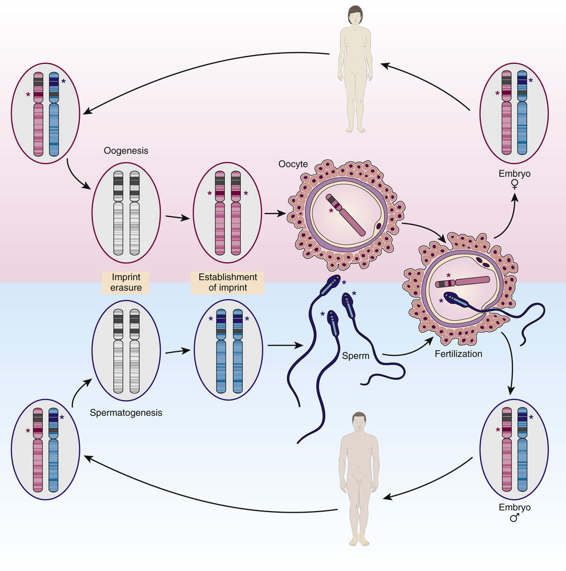
Imprinting takes place during gametogenesis, before fertilization, and marks certain genes as having come from the mother or father (Fig. 3-12). After conception, the parent-of-origin imprint is maintained in some or all of the somatic tissues of the embryo and silences gene expression on allele(s) within the imprinted region; whereas some imprinted genes show monoallelic expression throughout the embryo, others show tissue-specific imprinting, especially in the placenta, with biallelic expression in other tissues. The imprinted state persists postnatally into adulthood through hundreds of cell divisions so that only the maternal or paternal copy of the gene is expressed. Yet, imprinting must be reversible: a paternally derived allele, when it is inherited by a female, must be converted in her germline so that she can then pass it on with a maternal imprint to her offspring. Likewise, an imprinted maternally derived allele, when it is inherited by a male, must be converted in his germline so that he can pass it on as a paternally imprinted allele to his offspring (see Fig. 3-12). Control over this conversion process appears to be governed by specific DNA elements called imprinting control regions or imprinting centers that are located within imprinted regions throughout the genome; although their precise mechanism of action is not known, many appear to involve ncRNAs that initiate the epigenetic change in chromatin, which then spreads outward along the chromosome over the imprinted region. Notably, although the imprinted region can encompass more than a single gene, this form of monoallelic expression is confined to a delimited genomic segment, typically a few hundred kilobase pairs to a few megabases in overall size; this distinguishes genomic imprinting both from the more general form of random monoallelic expression described earlier (which appears to involve individual genes under locus-specific control) and from X chromosome inactivation, described in the next section (which involves genes along the entire chromosome).

Stay updated, free articles. Join our Telegram channel

Full access? Get Clinical Tree