Chapter 12 Gene Expression
I. RNA transcription
A. Overview
1. RNA is a single-stranded molecule that often contains internal base-paired regions, forming stem loops.
3. Prokaryotic mRNA is synthesized as a final product, but eukaryotic mRNA is synthesized as a primary transcript that must be further processed to remove introns.
B. Types of RNA
1. The three major types of RNA have specific functions in protein synthesis.
a. Messenger RNAs (mRNAs) provide the blueprint for an amino acid sequence; mRNA is about 5% of total RNA.
b. Transfer RNAs (tRNAs) match genetic information to an amino acid sequence; tRNA is about 15% of total RNA.
C. RNA polymerase
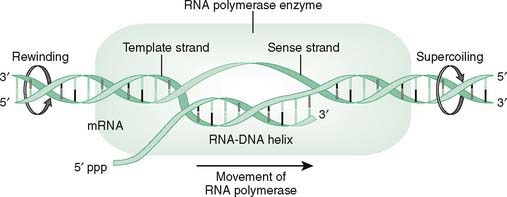
1. RNA polymerase unwinds the DNA helix and transcribes the DNA template strand into RNA, beginning at the transcription start site.
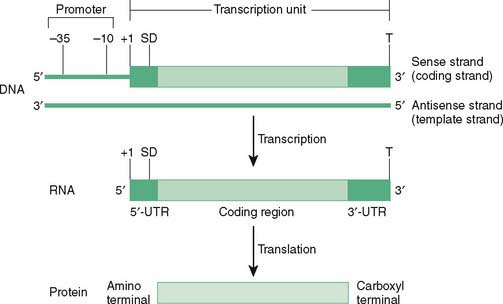
D. Prokaryotic transcription (Fig. 12-1)
1. Prokaryotic genes are organized as transcription units preceded by regulatory sequences called promoters.
2. The core enzyme, RNA polymerase, binds first to the promoter region in a precise orientation and then begins synthesis of RNA.
3. A transcription bubble (Fig. 12-2) is created as the helix opens for RNA polymerase transcription with closure of the bubble as the DNA reassociates.
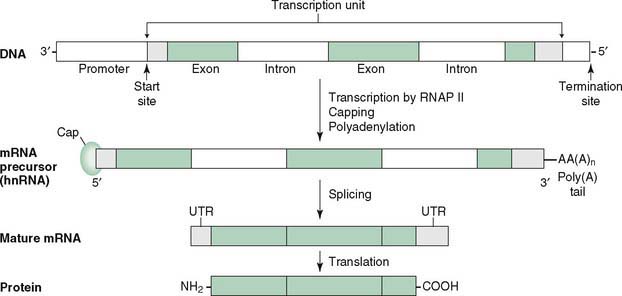
E. Eukaryotic transcription of mRNA (Fig. 12-3)
1. mRNA synthesis in eukaryotes begins with the binding of RNA polymerase II at specific DNA sequences known as promoters.
2. The initiation complex, containing RNA polymerase II and several basal transcription factors that bind RNA polymerase II, is assembled near the transcription start site (Fig. 12-4).
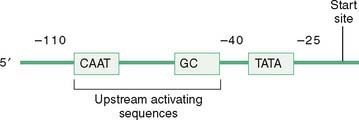
a. The TATA box, a conserved promoter sequence about 25 bases upstream of the start site, helps orient the RNA polymerase in many eukaryotic genes.
b. The CAAT box, another conserved sequence within the promoter region, is often present about 70 bases upstream of the start site.
3. Elongation of the RNA strand by RNA polymerase II continues until the enzyme reaches a termination signal.
F. Processing of the primary mRNA transcript
1. Processing of hnRNA occurs in the nucleus, yielding functional mRNA, which is exported to the cytosol for translation (see Fig. 12-3).
2. A 7-methylguanosine cap is added to the 5′ end of the growing mRNA before transcription is completed.
3. Polyadenylation at the poly(A) site leads to formation of the poly(A) tail consisting of 20 to 250 adenylate residues at the 3′ end of the transcript.
4. Splicing removes introns from capped, tailed transcripts and rejoins exons in a continuous coding sequence.
a. Splice sites, the junctions between exons and introns, usually are marked by consensus sequences: GU at the 5′ end of an intron and AG at the 3′ end.
II. Transcriptional Control of Gene Expression
A. Overview
1. The prokaryotic lac operon demonstrates positive control (i.e., factors required to initiate RNA synthesis) and negative control (i.e., factors required to prevent initiation of RNA synthesis).
2. Eukaryotic gene expression is regulated by controlling physical access to the DNA by the RNA polymerase and by controlling the rate of its binding.
4. Some exons can be spliced in or spliced out as a way of including or excluding domains; the final product is same gene product with different properties (e.g., membrane bound or soluble).
5. Editing of the final mRNA allows elimination of domains from carboxyl end of protein if the editing inserts a termination codon.
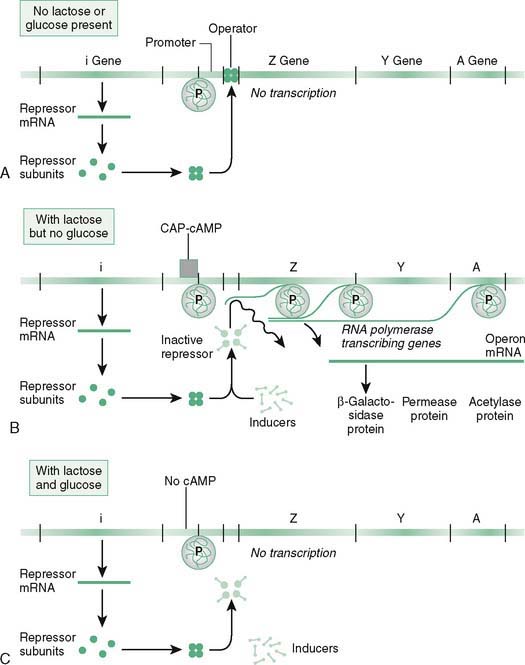
B. Prokaryotic control of gene expression (Fig. 12-5)
1. Regulation of the lac operon illustrates several modes of regulation of RNA transcription in bacteria:
a. Negative control occurs when only glucose is available as an energy source and expression of the lac operon is not needed.
b. Positive control occurs when lactose is present (with or without glucose present) and expression of the lac operon is needed.
(1) If lactose is present, the inducer, allolactose, is formed followed by binding the repressor to inactivate it.
(2) If no glucose is present, cAMP is elevated and combines with the catabolite activator protein (CAP), which binds to the promoter stimulating binding of RNA polymerase.
C. Eukaryotic control of gene expression
1. Eukaryotic gene expression is regulated by controlling physical access to the DNA by the RNA polymerase and by controlling the rate of its binding.
2. Physical accessibility of DNA to transcription factors and RNA polymerase II depends on the degree of compaction of chromatin in the region of a gene.
Heterochromatin: highly condensed, DNase resistant, highly methylated, and inactive
Euchromatin: dispersed, DNase digestible, highly acetylated, and active
a. Heterochromatin is highly condensed, DNase resistant, and inactive in transcription; the DNA has CpG islands that are highly methylated.
3. The rate of binding RNA polymerase to the promoter is regulated by transcription factors that bind to special DNA sequences.
a. Sequences in the promoter that bind transcription factors are those mentioned above (see Section I, E, 2): the CAAT box, the TATA box, and the CG box (the CG box is not the same as CpG islands).
b. Other regulatory elements in the DNA sequence, called enhancers or silencers, bind DNA regulatory proteins (i.e., specific transcription factors) that may activate or repress transcription by RNA polymerase II.




Stay updated, free articles. Join our Telegram channel

Full access? Get Clinical Tree