It is this mechanistic association with segmental duplications that distinguishes this subgroup of deletion and duplication syndromes from others whose breakpoints are highly variable and are not associated with any identifiable genomic feature(s), and whose mechanistic basis appears idiopathic (see Table 6-1). Here we focus on syndromes involving chromosome 22 to illustrate underlying genomic features of this class of disorders.
Deletions and Duplications Involving Chromosome 22q11.2
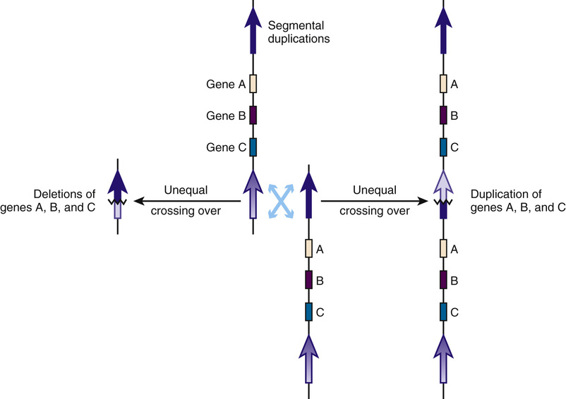
Several deletions and duplications mediated by unequal recombination between segmental duplications have been documented within the proximal long arm of chromosome 22 and illustrate the general concept of genomic disorders (Fig. 6-4). A particularly common microdeletion involves chromosome 22q11.2 and is associated with diagnoses of DiGeorge syndrome, velocardiofacial syndrome, and conotruncal anomaly face syndrome. All three clinical syndromes are autosomal conditions with variable clinical expression, caused by a deletion of approximately 3 Mb within 22q11.2 on one copy of chromosome 22. The microdeletion and other rearrangements of this region shown in Figure 6-5 are each mediated by homologous recombination between segmental duplications in the region. The deletions are detected in approximately 1 in 4000 live births, making this one of the most common genomic rearrangements associated with important clinical phenotypes.

Stay updated, free articles. Join our Telegram channel

Full access? Get Clinical Tree


