I. INTRODUCTION. Clinical molecular diagnostic methods have been integrated into many laboratory disciplines, and guidelines and recommendations from both professional societies and regulatory agencies have been developed to assist in the development and performance of clinical molecular pathology testing (Table 60.1). Most molecular tests performed in surgical pathology focus on somatic or acquired DNA variations in the cells of the disease process that provide information that aids in diagnosis, identifies prognostic indicators, stratifies patients into effective treatment options, helps monitor treatment response, and/or identifies patients at increased risk of disease. Because polymerase chain reaction (PCR)-based approaches are quick, reliable, and sensitive, PCR has become a central technology for much of clinical molecular genetic testing.
II. SPECIMEN REQUIREMENTS, HANDLING, AND PROCESSING. Clinical PCR-based molecular testing in the setting of surgical pathology requires the same attention to detail regarding efficient specimen collection, identification, preparation, and routing as any other pathology test.
A. Specimens. Specimen requirements are dictated by the disease process, including the type of tissue, amount of tissue, type of sample (fresh or frozen tissue, formalin-fixed paraffin-embedded tissue [FFPE], cytology specimen, and so on), and the extent of the disease in the sample. The amount of tissue required for PCR-based testing is relatively small, which contributes to the clinical utility of the technique.
Regardless of specimen type, two general features of the tissue sample influence molecular assays. First, there must be a sufficient quantity of the specific target cell (and therefore target DNA or RNA) in the sample. Second, the size or integrity of the nucleic acid molecules after isolation from the tissue can dramatically affect the sensitivity of the detection of specific alterations, thus nucleic acid degradation (whether due to fixation, or enzymatic, heat, pH, or mechanical forces) can reduce the sensitivity of testing.
1. Tissue type. Fresh peripheral blood, bone marrow, solid tissue biopsies, cytology specimens, enriched cell populations (e.g., from flow cytometry), and FFPE tissue sections are all sources of nucleic acids for molecular analysis. Specimens should be collected and transported to the molecular pathology laboratory using aseptic techniques, if possible. Transport on ice reduces cell lysis, minimizes nuclease activity, and reduces nucleic acid degradation.
2. Tissue quality
a. Fresh tissue and cell suspensions are the optimal templates for PCR. The preferred method of preservation of fresh tissue prior to isolation of nucleic acids is ultra-low-temperature frozen storage at −70°C, which permits indefinite preservation with virtually no effect on the quality of the extracted nucleic acids. Low-temperature frozen storage around −20°C can adequately preserve DNA and RNA for several months.
Cell suspensions (including hematologic specimens such as peripheral blood and bone marrow) should be collected in the presence of an anticoagulant, preferably ethylenediaminetetraacetic acid (EDTA) or acid citrate dextrose (ACD); heparin should be avoided because heparin carryover after nucleic acid isolation may inhibit subsequent PCR steps. Freezing of hematologic specimens presents distinct obstacles to the preparation of good quality nucleic acid and should generally be avoided.
TABLE 60.1 Selected Resources for Clinical Molecular Pathology Laboratory Operational Guidelines
Entity
Site
Tool(s)
Clinical Laboratory Improvement Amendments ’88
http://www.cms.hhs.gov/clia/
Centers for Medicare and Medicaid Services
Clinical Laboratory Accreditation requirements and compliance lists
57 Federal Register 7137-7186 (1992)
College of American Pathologists (CAP)
http://www.cap.org
Laboratory Accreditation (LAP)
Molecular Pathology Laboratory Inspection Checklist
Proficiency surveys
Molecular oncology (MO)
Medical genetics (MGL)
Pharmacogenetics (PGX)
Monitoring engraftment (ME)
Molecular microbiology (HIV, HCV, ID)
Microsatellite instability (MSI)
Sarcoma translocation (SARC)
Nucleic acid testing (viral; NAT)
Clinical and Laboratory Standards Institute (CLSI; formerly National Committee for Clinical Laboratory Standards, NCCLS)
http://www.nccls.org
Molecular Methods Guidelines
Genetic diseases
IGH & TCR gene rearrangements
Nucleic acid amplification
Nucleic acid sequencing
Collection and handling of specimens
Proficiency testing
American College of Medical Genetics (ACMG)
http://www.acmg.net
Standards and Guidelines for Clinical Genetics Laboratories
Policy Statements for Molecular testing of genetic diseases
US Food and Drug Administration (FDA)
http://www.fda.gov
Medical Devices 21CFR809.30
In Vitro Diagnostic Products for Human Use
Analyte-specific reagents (ASRs)
Association for Molecular Pathology (AMP)
http://www.amp.org
Molecular Pathology professional organization (incl. genetics, hematopathology, infectious disease, solid tumors)
CHAMP listserve for AMP members
Test directories
Solid tumors
Hematopathology
Infectious disease
b. Nucleic acids extracted from fixed tissue can also be used in PCR, although the type of fixative and length of fixation both have a profound effect on their recovery. Non-cross-linking fixatives such as ethanol provide the most consistent preservation of amplifiable DNA, with more variability from tissues fixed with formalin, Zamboni’s and Clark’s fixatives, paraformaldehyde, and formalin-alcohol-acetic acid. Tissues processed in Carnoy’s, Zenker’s, Bouin’s and B-5 fixatives are poor substrates for PCR testing since little amplifiable DNA can be recovered from them.
The effects of formalin fixation on PCR-based testing have been evaluated in some detail, not surprising given that most surgical specimens are fixed in formalin. Formaldehyde reacts with nucleic acids and proteins to form a mixture of end products that are covalently linked by methylene bridges. Thus, the quality of DNA isolated from formalin-fixed tissue is critically dependent on the length of fixation, with a deterioration of PCR signal with increasing fixation time. In general, tissue fixed in neutral buffered formalin for <8 hours contains DNA and RNA from which PCR products >600 bp in length can be reliably amplified, but fixation extended for greater than 8 to 12 hours decreases the length of PCR products that can consistently be amplified.
3. Tissue quantity. Minimum sample requirements are determined by the assay methodology and extent of target cell involvement in the tissue. A typical PCR-based assay requires only 20 to 200 ng of DNA (about 103 to 104 cells), although multiplexed PCR may require a bit more DNA in order to equally represent all targets. The sensitivity of PCR for detection of a few target molecules in a large background of unaltered DNA molecules (1 in 105) is one of the principle strengths of the methodology.
III. ANALYTIC/TECHNICAL VERSUS DIAGNOSTIC AND OPERATIONAL ASPECTS OF TESTING. The familiar probabilistic model used to define the likelihood that a particular patient is correctly classified on the basis of a test result (Table 60.2) can be applied to molecular genetic tests as with any other laboratory test. However, it is important to recognize that the quantitative performance of a lab test can be evaluated at four different levels (Table 60.3), and that test performance at one of the four levels does not necessarily predict performance at the other levels. Differences between these four levels of analysis are often overlooked even though they account for many of the confusing or seemingly conflicting results regarding the utility of molecular genetic testing in surgical pathology.
IV. BASIC PCR METHODOLOGY
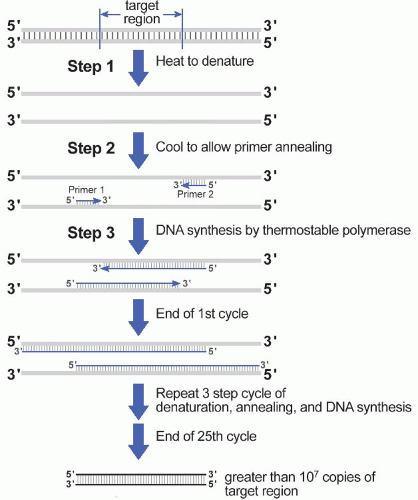
A. Amplification. Selective amplification of the target sequence is achieved through the use of oligonucleotide primers that hybridize to the 5′ and 3′ ends of the DNA target sequence (Fig. 60.1). In addition to the two primers and input (template) DNA, the reaction mixture also includes the four deoxynucleotide triphosphates (dATP, dCTP, dGTP, dTTP) and a heat-stable (thermostable) DNA polymerase. The first step of the PCR itself involves heating the mixture to a high temperature to denature the target DNA; in the second step, the reaction is cooled to allow the primers to anneal to their complementary sequence in the target DNA; in the third step, the reaction is heated to the temperature at which the heat-stable DNA polymerase has optimal activity. As a result of this three step denaturation, annealing, and polymerization cycle, the two primers will initiate synthesis of new DNA molecules from opposite strands of the input DNA heteroduplex. With each repetition of the three-step cycle, the newly synthesized DNA strands
will also act as templates for further DNA synthesis, and so DNA duplexes in which both strands have the fixed length of the target sequence (so-called amplicons) accumulate exponentially.
TABLE 60.2 Nomenclature When Bayes’ Theorem for One Variate Is Applied in Laboratory Testing
Number of subjects with positive test result
Number of subjects with negative test result
Number of subjects with disease
TP
FN
Number of subjects without disease
FP
TN
TP, True positives or number of diseased patients correctly classified by the test; FP, false positives or number of patients without the disease misclassified by the test; FN, false negatives or number of diseased patients misclassified by the test; TN, true negatives or number of patients without the disease correctly classified by the test.
Diagnostic sensitivity =
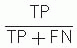
Diagnostic specificity =
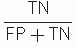
Predictive value of positive test =
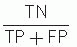
Predictive value of negative test =
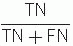
Efficiency of the test or number fraction of patients correctly classified =
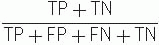 Youden index = [(sensitivity + specificity) − 1]
Youden index = [(sensitivity + specificity) − 1]
PCR makes it possible to selectively amplify a specific DNA target sequence within a background of heterogeneous DNA sequences, such as total genomic DNA or cDNA derived from unfractionated cellular RNA. However, each of the components in a PCR, including the input DNA, the oligonucleotide primers, the thermostable polymerase, the buffer, and the cycle parameters, has an effect on the sensitivity, specificity, and fidelity of the reaction.
B. Factors that affect PCR testing on an analytic/technical level
1. Advantages of PCR
a. PCR is simple, quick, and inexpensive. A single PCR cycle of melting, annealing, and extension is usually completed within several minutes, and consequently an entire PCR amplification of 25 to 35 cycles can be performed in only a few hours. Because of the high level of amplification achieved by PCR, the product DNA can be visualized after simple gel electrophoresis, avoiding the hazards and expense of radiolabeling methods.
TABLE 60.3 Four Levels at Which a Laboratory Test Can Be Evaluated
Level
Measures
Analytic/technical
Technical sensitivity, technical specificity, precision, accuracy, proficiency
Diagnostic
Diagnostic sensitivity, diagnostic specificity, Youden index
Operational
Predictive value of a positive result, predictive value of a negative result, efficiency
Medical decision making
Cost-benefit analysis, stratification to optimal treatment regimen, stratification to gene-targeted treatment regimen
b. PCR has high sensitivity and specificity. When optimized, PCR can detect one abnormal cell in a background of 105 normal cells, and can even be used to analyze single copy genes from individual cells (Methods Enzymol. 2002;356:295, 334). PCR can also be used to detect a broad range of genetic abnormalities ranging from gross structural alterations such as translocations to single base-pair changes.
c. PCR products are easily labeled for detection. For primer-mediated labeling, a labeled chemical group (usually a fluorophore) is attached to the 5′ end of either or both oligonucleotide primers. Alternatively, the PCR product can be directly labeled by including one or more labeled nucleotide precursors into the PCR mix.
d. Phenotype-genotype correlations are possible. When performed on tissue sections, PCR provides only an indirect correlation of morphology with underlying genetic abnormalities. Microdissection, in which the region of interest is carved out of the FFPE tissue block, scraped from tissue sections or cytology slides, or collected more precisely with a micromanipulator apparatus, provides some enrichment for morphologic-genetic correlations. More precise phenotypic-genotypic analysis is achieved by collecting individual cells by laser capture microdissection, by flow cytometry, or even by immunomagnetic methods. In situ PCR performed on histologic tissue sections themselves is perhaps the ultimate method for providing morphologic localization of genotypic expression; however, the technique is so technically demanding that it has limited use in clinical laboratories.
2. Limitations of PCR
a. PCR only analyzes the target region. Testing only provides information on the target segment amplified by the specific primer set employed.
b. PCR only amplifies intact target regions. Mutations that damage a primer binding site (including insertions, deletions, and even point mutations) preclude amplification of the target region by PCR and can easily lead to errors in test interpretation. Similarly, mutations that alter the structure of the target region in ways not accounted for during primer set design (e.g., large insertions, deletions, inversions, or translocations) may preclude amplification.
Stay updated, free articles. Join our Telegram channel

Full access? Get Clinical Tree


Direct and Indirect Methods for DNA Sequence Analysis
Direct and Indirect Methods for DNA Sequence Analysis
TuDong Nguyen
Barbara Zehnbauer
John D. Pfeifer