CASE STUDY
A 30-year-old man developed liver cirrhosis and diabetes. Laboratory studies showing high serum ferritin and high transferrin saturation led to the hypothesis that iron toxicity to the liver and pancreas contributed to his cirrhosis and diabetes. A blood specimen was tested for mutation in the HFE gene that is responsible, at least in part, for hereditary hemochromatosis. Polymerase chain reaction (PCR) followed by melt curve analysis was performed, and a pathologist interpreted the findings as HFE C282Y mutation without wild-type DNA at that locus. Homozygous HFE gene mutation with the predicted amino acid substitution predisposes to iron overload by overabsorption of iron from the diet. He was treated with therapeutic phlebotomy until his serum iron levels returned to the normal range. He remains at risk for iron overload, and he should be periodically monitored and managed accordingly. A genetic counselor educated him about the increased risk of iron overload faced by blood relatives if they, too, inherited two mutated alleles of the HFE gene.
The practice of molecular pathology capitalizes on analysis of deoxyribonucleic acid (DNA) and ribonucleic acid (RNA) to inform medical decision making. Each nucleated cell in the body contains a complete set of DNA inherited from parents, constituting the person’s genome. DNA is the blueprint by which cells catalog, express, and transmit genetic information. The human genome is composed of 3 billion pairs of nucleotides divided among 46 chromosomes containing about 25,000 different protein-coding genes plus additional genes encoding RNA that is not translated into protein. There are two copies of every gene—one of paternal and the other of maternal origin. Transcription factors act on gene promoters to regulate gene transcription, as do chromatin modifications such as DNA methylation and histone acetylation.
MOLECULAR TECHNOLOGIES
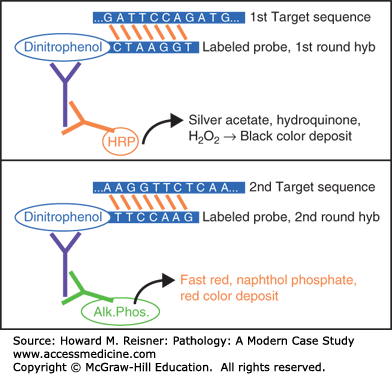
Molecular assays rely on the ability to find a specific nucleotide sequence in DNA or RNA by using a nucleic acid probe targeting that sequence. A probe is a single-stranded segment of nucleic acid whose nucleotide sequence is complementary to the target sequence of interest. A probe binds to its target through a process called hybridization, and then the probe is detected or its effects (e.g., priming DNA synthesis) are evaluated using various detection strategies (see Figure 5-1).
FIGURE 5-1
DNA is composed of complementary strands of nucleotides that are connected by hydrogen bonds between adenine (A) and thymine (T), or between guanine (G) and cytosine (C). In the laboratory, the two strands of DNA may be dissociated from one another by heating them to near boiling (94°C) or by treating with an alkaline solution (high pH). Single-stranded patient nucleic acid may then hybridize to a complementary probe. In the examples depicted here, labeled probes hybridize to target sequences in patient DNA, and then a detection system deposits black or red chromagen at the locus where the probe is bound.
QUICK REVIEW
The molecular technologies most commonly implemented in clinical settings are PCR, in situ hybridization, DNA sequencing, Southern blot analysis, and array technology. Benchwork is performed by medical technologists, while pathologists and other clinical laboratory scientists are responsible for interpreting findings and for consulting with health-care providers on the indications of testing and the medical implications of results.
MEDICAL APPLICATIONS OF MOLECULAR TECHNOLOGY
Analysis of nucleic acid sequences in patient samples helps in diagnosis, classification, prognosis, predicting efficacy of therapy, documenting response to therapy, monitoring disease burden, identifying relapse, and screening for disease in high-risk populations. The range of diseases is quite broad and includes heritable disease, cancer, and infectious disease. Heritable disease is, by definition, a disorder resulting from abnormal DNA (chromosomal DNA or mitochondrial DNA). Cancers harbor acquired genetic changes that are responsible for malignant behavior, such as an activated oncogene or an inactivated tumor suppressor gene driving cell proliferation. Finding a tumor-related molecular defect can help confirm a diagnosis of cancer and can further serve as a tumor marker for monitoring disease burden during therapy. For infectious disease diagnosis, each pathogen has its own genome composed of DNA or RNA for which molecular assays may indicate the presence of, amount of, and drug-resistance sequences encoded by a given microbial agent. Drug efficacy or toxicity might be influenced by sequence variants encoding factors important in drug metabolism, distribution, or effectiveness. A pharmacogenetic test can predict which drug or dose of drug is most likely to improve outcome in a given patient.
Specimens submitted to a clinical laboratory for genetic testing are collected and handled with care to preserve intact DNA and RNA for downstream analysis. DNA and RNA can be extracted from a wide range of specimens (e.g., blood, marrow and other body fluids, frozen or formalin fixed paraffin embedded tissue, buccal swabs, and needle aspirates) by first using detergent and proteinase enzymes to lyse cellular lipid membranes and degrade proteins. Semiautomated instruments facilitate further purification of DNA or RNA based on preferential binding to a glass or similar surface. DNA and RNA are quantified by spectrophotometry. While DNA is relatively stable, RNA is unstable by virtue of its susceptibility to degradation by ubiquitous enzymes collectively termed RNase. Special precautions are required to assure the integrity and quality of nucleic acid from the time of collection through the various hybridization procedures described below. Instructions for collection, transport, and storage of specimens are provided by the testing laboratory.
LABORATORY PROCEDURES
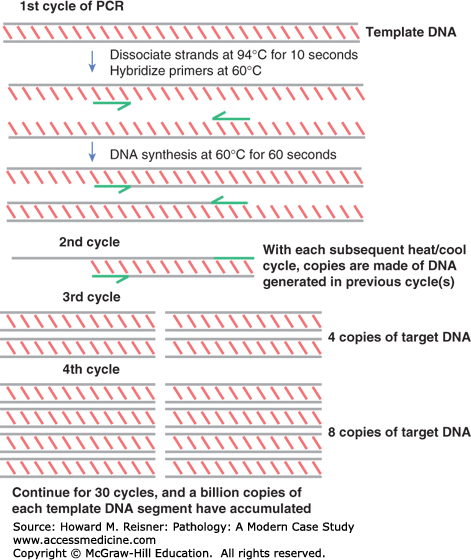
PCR is a method of replicating a short sequence of DNA, typically about 100 bases in length, so that it may be more easily detected, quantified, or further analyzed for genetic alterations. Although several other technologies have been invented to amplify nucleic acid sequences or probe signals, PCR remains the most commonly used analytic method in molecular testing laboratories (see Figure 5-2).
FIGURE 5-2
PCR is a method of enzymatically amplifying a particular segment of DNA through a process of repeated cycles of heating, cooling, and DNA synthesis in an instrument called a thermocycler. First, patient DNA is mixed with two short oligonucleotide probes called primers (green half-arrows) that define the segment to be amplified. Also in the mix is an enzyme called DNA polymerase that begins at the 3′ end of the primer and proceeds to incorporate nucleotides to make double-stranded DNA. In each cycle of the reaction, the sample is first heated to dissociate the two strands, then cooled to facilitate primer binding and initiation of enzymatic DNA replication. In each cycle, the products of previous cycles serve as templates, permitting exponential accumulation of replicate DNA copies. After 30 cycles, which takes only a couple of hours, up to 230 (about a billion) copies of the DNA segment have been generated. Depending on the purpose of the test, the PCR products are then detected, quantified, or further analyzed for sequence variants.
PCR is the technology of choice among pathologists because of (1) exquisite sensitivity to low numbers of target sequences, (2) small sample size requirement (theoretically only one molecule), and (3) speedy and relatively inexpensive automated test systems. The major disadvantage is the extreme care required to avoid contamination of samples and reagents by extraneous DNA. Abundant amplicons generated from previous reactions are especially concerning, and strategies have been developed to fragment these potential contaminants. Physical separation of pre- and postamplification work areas is recommended.
Interestingly, the DNA polymerase used in PCR reactions is “thermostable,” meaning that it can endure near-boiling temperatures and still remain active. This feature allows the reaction to continue during repeated cycles of heating and cooling without significant loss of enzymatic function. The polymerase is derived from bacteria such as Thermus aquaticus (Taq) that evolutionarily adapted to live around geysers or hot springs.
In real-time PCR, one or two fluorescent internal probes are added to the mixture so that they may mark amplified products to be quantified by a sensor during every amplification cycle. By comparing results to a series of standards, one can extrapolate how much target DNA was present in the original sample. This procedure may also be called quantitative PCR (see Figure 5-3).
FIGURE 5-3
Real-time PCR is a technology to precisely measure amount of DNA template in a patient sample by monitoring the amount of fluorescence at the end of each amplification cycle in comparison to a series of standards of known DNA level.
Stay updated, free articles. Join our Telegram channel

Full access? Get Clinical Tree